Robust Amplicon recovery post-Crystal Digital PCR™ for downstream genomic applications
Introduction
Droplet microfluidics technologies, and particularly digital PCR (dPCR), have provided highly precise and sensitive detection of genetic targets. There is often an imperative for researchers to use samples post PCR for downstream applications due to their scarcity. In this scenario, there is a need for methods that limit the required input volume of a scarce sample and allow the integration of multiple sequential downstream workflows to promote full characterization and validation of experimental findings. Historically, recovery of digital PCR products for downstream genomic testing applications has been challenging due to limitations imposed by the digital PCR technology. Stilla Technologies has developed Crystal Digital PCR™ (cdPCR) on the naica® system, a flexible and high multiplex droplet fluorescence imaging based dPCR platform that allows for multi-color detection of genetic targets, including the ability to recover amplified targets (Amplicons) after cdPCR is complete.
This study highlights the Geode instrument, which performs the droplet partitioning and thermocycling of the cdPCR workflow, and has a simple dedicated command incorporated for direct amplicon recovery from the analyzed droplets. After imaging the chip, the amplicons contained in the recovered emulsion are extracted using a standard chloroform protocol that takes less than one hour. Recovered DNA from the Droplet Crystal is compatible with a variety of downstream molecular appli- cations including but not limited to Sanger Sequencing, Next Generation Sequencing (NGS) and additional rounds of cdPCR.
Post-cdPCR Amplicon recovery is an excellent tool to support orthogonal validation of cdPCR results using other genomic methods. Furthermore, if combined with additional technologies such as sequencing, amplicon recovery provides discovery capabilities of additional sequence anomalies (eg point mutations, Single Nucleotide Polymorphism (SNPs) and insertions or deletions contained within the amplicons not targeted in the original assay). Here, this study presents the naica® system amplicon recovery protocol and shows how it can be seamlessly combined with cdPCR reamplification, Sanger Sequencing and NGS using the Illumina TruSight™ Tumor 15 kit.
Amplicon recovery protocol
Amplicon recovery is available on the naica® system for the Sapphire Chip. Upon completion of the partitioning and amplification in the Geode instrument, the Droplet Crystal is imaged on the Prism3 or Prism6 instrument. Once the scanning is complete, the Sapphire Chip is then placed back in the Geode instrument and the amplicon recovery program is executed (Figure 1, step 1). Following, to recover the droplet emulsion, the blue caps of the Sapphire Chip are removed and all contents of the well (40-50 ul) are transferred with a pipet (Figure 1, step 2) to a microcentrifuge tube (Figure 1, step 3).

The emulsion is allowed to separate on the bench for 1-2 minutes, and the bottom oil phase is removed from the tube and discarded. The DNA is then extracted by adding 20 µL of TE and 70 µL of chloroform to the remaining aqueous phase. The solution is mixed by vortexing and centrifuged at high speed for 10 minutes to separate the aqueous and organic phases. The upper aqueous phase containing the DNA is then transferred by pipetting to a clean microcentrifuge tube, taking care to avoid the interface between the aqueous and organic phases. The recovered amplicon is now ready to be used in downstream workflows.
Amplicon recovery percentage
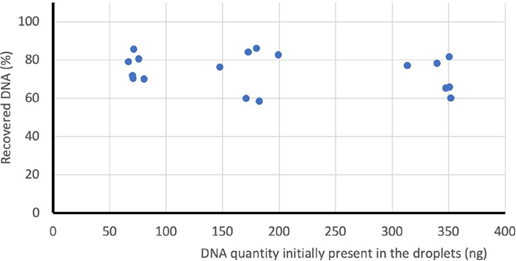
The percent recovery of amplicons was assessed by 1) performing cdPCR in Sapphire Chip on three known amounts of genomic DNA, 2) scanning the chips using the Prism3 instrument before and after the amplicon recovery protocol and using Crystal Miner software to determine the total number of droplets per reaction, 3) extracting amplicon DNA from the recovered droplet emulsion and 4) quantifying the amount of recovered amplicon by performing a second round of cdPCR (Figure 2). We observed that on average 98% of the droplets in the chamber were recovered from the Sapphire Chip and an average of 70% of the total DNA present in the initial Droplet Crystal was recovered (Figure 2).
To characterize the compatibility of recovered DNA amplicons with Sanger sequencing protocols, an 81 base pair amplicon of the human Albumin gene was recovered, and Sanger-sequenced by GATC (Eurofins Genomics, Cologne, Germany). Recovered amplicons were premixed with the forward or reverse primer and sequenced by two methods: SupremeRun and LightRun. Whereas the LightRun sequencing method permits the fastest time to results and is suitable for purified plasmids or PCR products, the SupremeRun method is optimized for difficult templates, such as GC-rich sequences, hairpins and secondary structures that often inhibit sequencing reactions. Both sequencing methods gave clean Sanger sequencing results with perfect alignment of forward and backward reads (data not shown), demonstrating high PCR product quality.
Recovering amplicons from a single positive droplet
We next evaluated the efficacy of amplicon recovery from Droplet Crystals having a low number of positive droplets. To this end, two targets were amplified by Crystal Digital PCR™ using low concentrations of human genomic DNA generating zero to four positive droplets per Sapphire Chip chamber. Application of the amplicon recovery protocol and subsequent Crystal Digital PCR™ of the recovered amplicons showed that DNA was successfully recovered from all Droplet Crystals initially containing at least one positive droplet, whereas no targeted DNA was detected in Droplet Crystals initially containing no template controls (NTC) (Table 1).
| Number of positive droplets in chamber | 0 (NTC) | 1 | 2 | 3 | 4 |
| Number of chambers with DNA recovered/ Total number of chambers tested | 0/3 | 4/4 | 9/9 | 4/4 | 3/3 |
Targeted NGS library preparation using Crystal Digital PCR™ and amplicon recovery
Next, we evaluated the compatibility of amplicons recovered after cdPCR with the downstream genomic application of NGS. NGS is a high-throughput approach for DNA sequencing and several platforms have been commercialized. In this study, Illumina® technology and the TruSight™ Tumor 15 kit were used to generate 250 amplicons from 15 cancer-associated genes: AKT1, BRAF, EGFR, ERBB2, FOXL2, GNA11, GNAQ, KIT, KRAS, MET,NRAS, PDGFRA, PIK3CA, RET and TP53, using two pools of primers: A and B. A commercially available wild-type human genomic DNA spiked with a known amount of commercially available mutant DNA accounting for 11 mutations was used as the library A and library B starting material. To evaluate the compatibility of cdPCR with NGS, we compared the techniques of cdPCR and in-tube bulk PCR for the construction of NGS libraries. The following two protocols were used to generate the final libraries to be sequenced: the first amplification step in library preparation (Step 1) was performed by either: 1) standard “in- tube” bulk PCR using a thermocycler, or 2) cdPCR using Sapphire Chips followed by amplicon recovery (Figure 3).
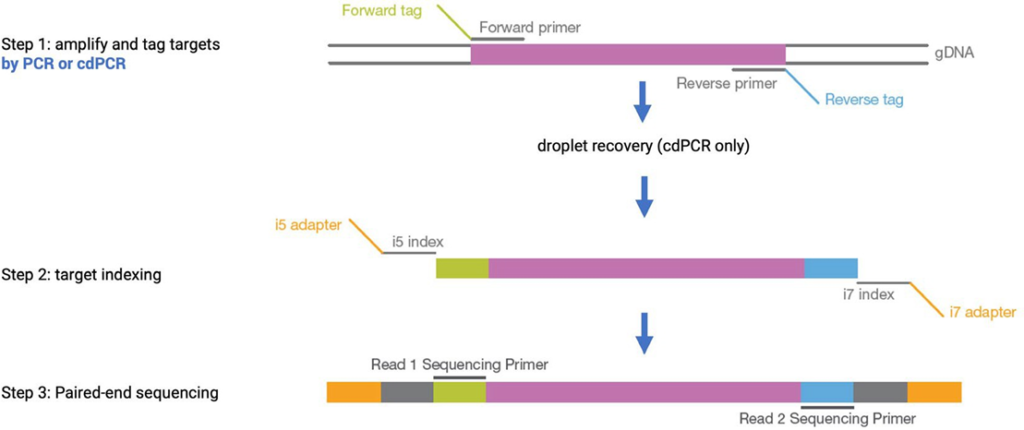
Both cdPCR and bulk PCR methods were used with primers from pool A and B of the TruSight™ kit to generate a total of eight barcoded libraries per PCR method and a total of 16 barcoded libraries. With each method two libraries from wild-type human genomic DNA (library A and library B) and six libraries from a mixture of wild-type and mutant DNA (triplicate of library A and triplicate of library B) were created (Figure 4).

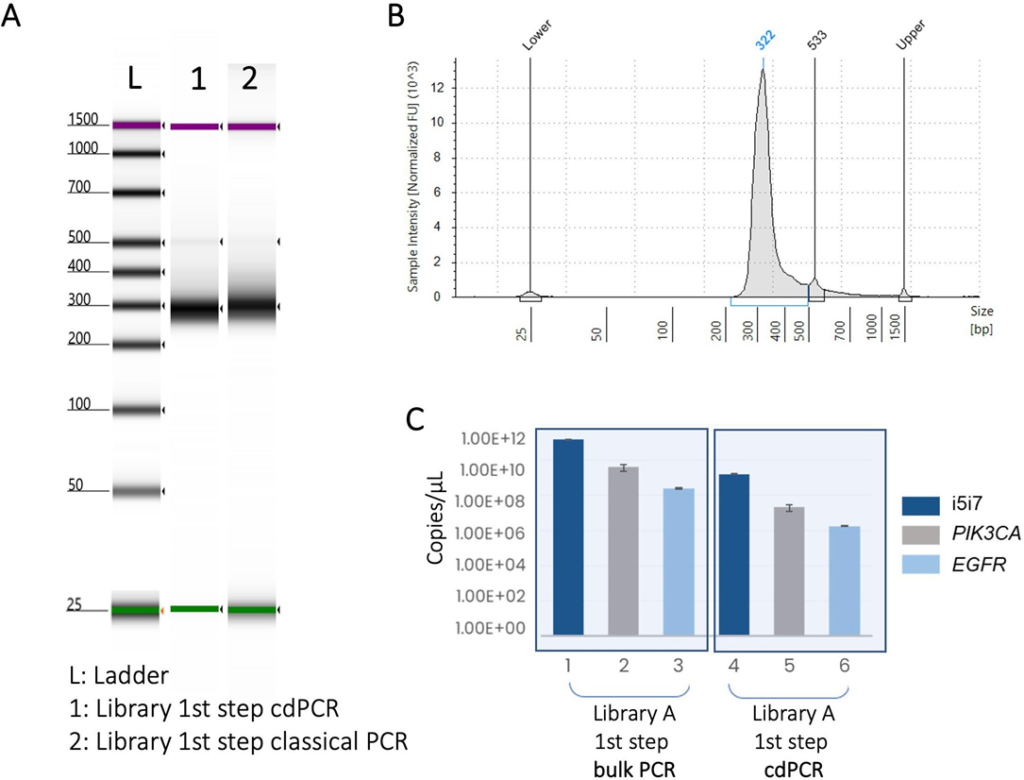
First, a qualitative quality control was performed on the DNA libraries created by bulk PCR and cdPCR using a 4200 TapeStation with the dsDNA ScreenTape (Agilent Technologies; Figure 5A and 5B). The size profiles of each library were highly similar for both preparation methods (Figure 5A) Next, cdPCR was used to perform a quantitative quality control on the DNA libraries generated by bulk PCR and cdPCR (Figure 5C). Briefly, three sequences (i5i7, PIK3CA, EGFR) present in the TruSight™ libraries were re-amplified by cdDNA from each library. In all cases, successful amplification was obtained (Figure 5C), showing that each library is of high quality and suitable for NGS.
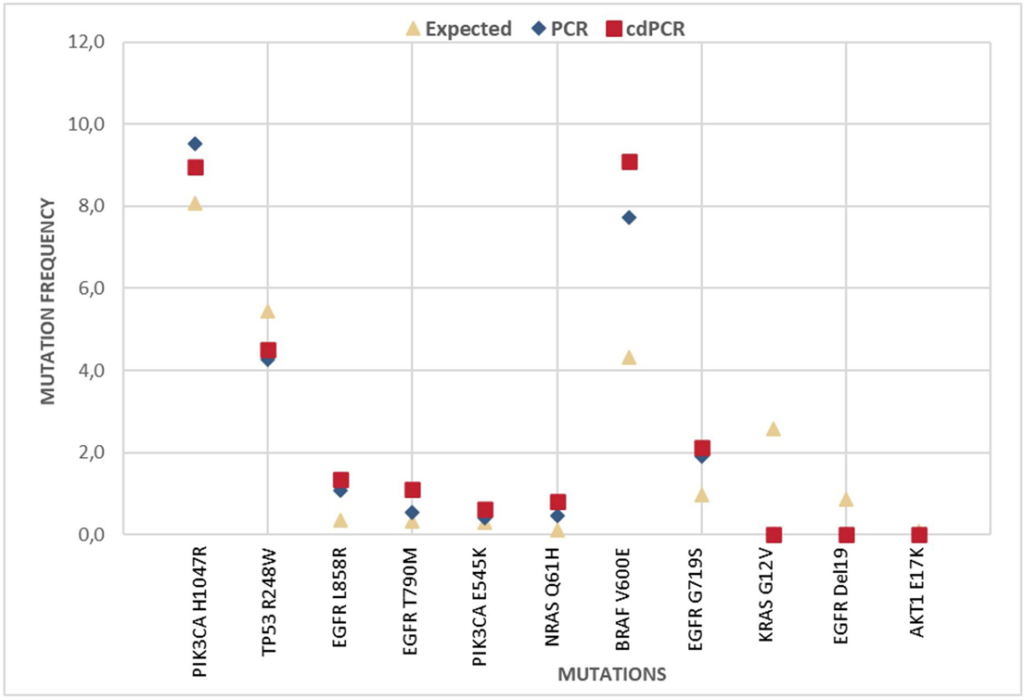
The eight libraries from the library preparation method (cdPCR or bulk PCR) were pooled together at equal amounts. Libraries were then sequenced on an Illumina® MiSeq (Integragen, Evry, France). Mutations were detected and mutation frequencies were calculated by dividing the mutant read count by depth position. The mutation frequencies detected in libraries created by bulk PCR and libraries created by cdPCR are reported and compared to the theoretical expected percentage (Figure 6).
The presented study demonstrates that amplicons recovered post-cdPCR on the naica® system are compatible with a variety of downstream genomics protocols, including but not limited to NGS and Sanger sequencing.
Technical Note Highlights
- Amplicons can be recovered from the analyzed Droplet Crystal post Crystal Digital PCR™ by a simple protocol available on the naica® system
- Up to 98% of droplets are recovered and an average of 70% of the total DNA present in the initial Droplet Crystal is recovered.
- Amplicons can be successfully recovered from a Droplet Crystal containing only one positive droplet.
- The cdPCR recovered amplicons can be Sanger- sequenced and reamplified by cdPCR.
- Crystal Digital PCR™ and amplicon recovery can be used to create targeted next generation sequencing libraries.
- The percentage of mutations sequenced from cdPCR-generated libraries is objectively accurate and correlates well to measurements from bulk PCR- prepared libraries.
Stilla thanks Jordan Madic who contributed to the adaptation of the next generation sequencing protocol for Crystal Digital PCR™.