Sensitive & flexible Crystal Digital PCR™ quantification using the naica® multiplex PCR MIX
A defining characteristic of digital Polymerase Chain Reaction (PCR) leading to its high sensitivity is the partitioning of the PCR reaction mix into thousands of individual compartments separated by an isolating phase. Chemical interactions occurring at the interface between the reaction mix and the isolating phase can dramatically impact PCR efficiency. The composition and performance of the PCR master mix are critical to ensure efficient PCR amplification of the targets and partition compatibility. The naica® multiplex PCR MIX was specially formulated to provide robust partition compatibility and excellent linear Crystal Digital PCR™ quantification of multiple nucleic acid targets across the entire dynamic range of the naica® system, even in the presence of a complex sample DNA background. To ensure the flexibility and sensitivity that our customers need, the naica® multiplex PCR MIX is available at both 5X and 10X concentrations, thus freeing up valuable reaction volume that can be instead occupied by additional sample input, probes and/or primers.
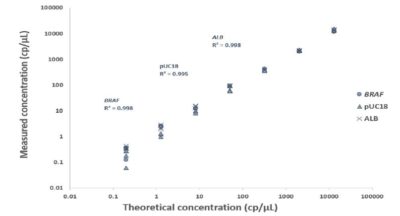
The naica® multiplex PCR MIX ensures simultaneous linear quantification of multiple targets across the dynamic range of the naica® system
The naica® multiplex PCR MIX was optimized for use on the naica® system to ensure compatibility with dual-labelled fluorescent probe multiplex assays. A stably incorporated blue channel reference dye ensures reliable partition detection and facilitates reaction assembly. Using the 10X naica® multiplex PCR MIX, simultaneous detection of three reference targets from 0.2 to 13,000 copies (cp)/µL was achieved on the naica® system (Figure 1). The high initial concentrations of the mix (available at both 5x and 10x concentrations) maximize the reaction volume available for sample input, thus increasing the potential to detect targets in dilute samples.
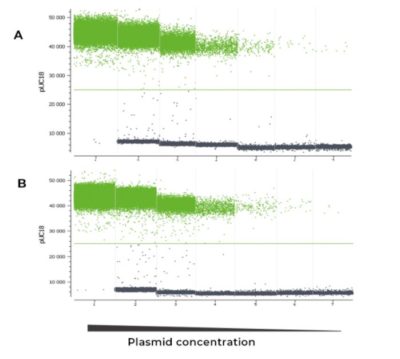
Stable and sensitive detection of a low concentration target in a multiplex assay
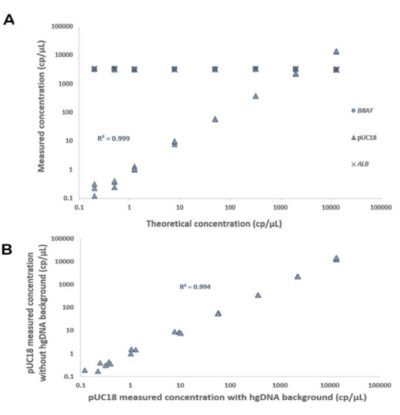
Co-amplification of several targets in a multiplex assay is more complex than a single amplification reaction due to potential crosstalk and reaction complexity, possibly reducing the linear range of detection. Using the naica® multiplex PCR MIX, the pUC18 target was equally and comparably quantified throughout the dynamic range of the naica® system using Sapphire chips in both a triplex assay (Figure 2A and 3A) in which both the BRAF and ALB hgDNA targets were present in high concentrations, and a simplex assay (Figure 2B and 3B) in which pUC18 template was the unique DNA present in the reaction. In addition, the fluorescence levels and separability of the pUC18 positive and negative populations were similar in the simplex and triplex reactions (Figure 2). Moreover, regardless of the simplex or triplex context, the pUC18 target was robustly and reliably quantified (Figure 3), and the detection range and the linearity of this detection were similar in the two contexts.
Technical Note Highlights
- The naica® multiplex PCR MIX enables an excellent linear Crystal Digital PCR™ quantification of multiple targets across the entire dynamic range of the naica® system.
- Using the naica® multiplex PCR MIX, a target is equally and comparably quantified in both a simplex and a triplex detection assay.
- naica® multiplex PCR MIX is available at both 5X and 10X concentrations, thus freeing up valuable reaction volume that can be instead occupied by additional sample input, probes and/or primers.