RNA Quantification with Crystal RT-dPCR
Multiplexing targets with Crystal RT-dPCR
Reverse-transcription PCR (RT-PCR) is widely used for RNA analysis, for exemple for genomic expression or viral detection, and is applied to many fields, from life science research to diagnostics. Crystal RT-dPCR enables straightforward multiplexing of targets (references, or transcripts of interest), absolute quantification of viral RNA without need for standard curves and allows exquisite detection of fine differences in gene expression.
Thanks to 3-color Crystal RT-dPCR, three different messenger RNA (mRNA) can be analyzed in a single one-step reaction, demonstrating that 1.5-fold differences in gene expression can easily and precisely be quantified (Figure 1).
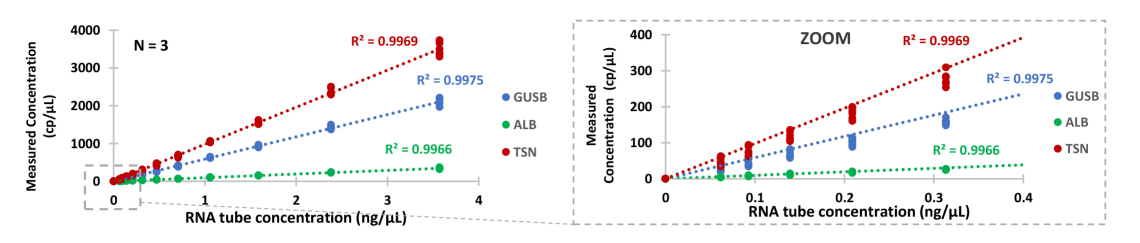
Figure 1 : Absolute quantification of mRNAs in total RNA using one-step Crystal RT-dPCR. Glucuronidase beta (GUSB), Albumin (ALB) and Translin (TSN) mRNAs were quantified by 3-color Crystal RT-dPCR in a serial dilution of total human RNA, ranging from 0.06 to 3.6 ng/μL with a ratio of 1.5 between each point. Assays were performed in Sapphire Chips using 1x qScript tm XLT One-Step RT-qPCR toughmix r , and FAM-, HEX- and Cy5-labeled TaqMan tm probes. Results for three replicates show good linearity, accuracy and reproducibility for the different levels of expression of each target. Slope of the regression line: GUSB = 588.8 cp/ng; ALB = 96.5 cp/ng; TSN = 980.6 cp/ng.
Quantification of low-level transcripts
Detecting low amounts of transcripts in a high background of total RNA can be challenging. Quantification with the naica® tm system shows excellent linearity and reproducibility for low fraction of transcript (Figure 2A). Reaction efficiency, as assessed through fluorescence amplitude, remains stable even in presence of a high load of total RNA (2μg – Figure 2B).
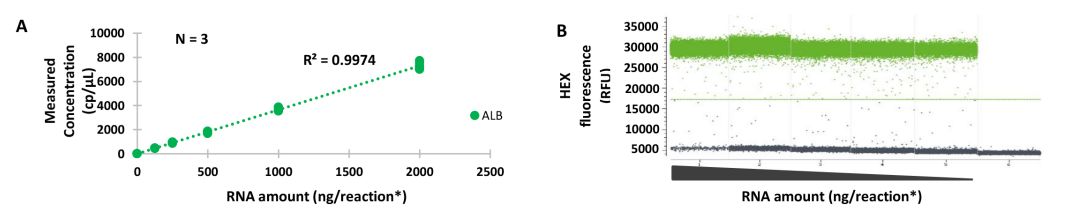
Figure 2 : Absolute quantification of mRNA with a high amount of total RNA using one-step Crystal RT-dPCR. Albumin (ALB) mRNA was quantified by Crystal RT-dPCR in a serial dilution of total human RNA, ranging from 2μg to 125 ng per reaction with a radio of 2 between each point. Assays were performed in Sapphire Chips using 1x qScript tm XLT One-Step RT-qPCR Toughmix r and HEX-labeled TaqMan tm probe. Linear plot shows accurate and reproducible results for the three duplicates (A), and no loss of efficiency has been observed on droplets fluorescence dotplots (B). RFU : Relative Fluorescence Units. *27 μL per reaction.
Viral RNA absolute quantification
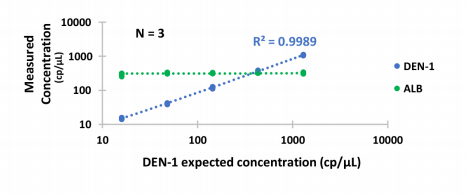
Dengue virus 1 (DEN-1) and its 3 other closely-related serotypes can cause fatal conditions, such as Dengue Hemorrhagic Fever and Dengue Shock Syndrome. No need for calibration curves using standards : in less than 2h30min, Crystal Digital RT-PCR enables viral quantification in a fast and user-friendly manner suitable to all labs and workflows.
Figure 3 : Absolute quantification of Dengue-1 RNA using one-step Crystal RT-dPCR. Dengue-1 RNA was quantified by Crystal RT-dPCR in a background of 300 cp of total human RNA, with ALB mRNA quantified as a reference. Assays were performed in Sapphire Chips using 1x qScript tm XLT One-Step RT-qPCR Toughmix r and FAM- and HEX-labeled TaqMan tm probes. Dengue-1 RNA was previously quantified by the manufacturer by digital PCR. Logarithmic plots show good concordance with the expected concentration and reproducible results.