On- and off-target detection in a CRISPR-edited mammalian cell line using a single-step 3-color Crystal Digital PCR™ assay
Stilla Technologies In collaboration with Moritz Kueblbeck, Andrea Callegari, Beatriz Serrano-Solano Dr. Jan Ellenberg Group European Molecular Biology Laboratory
Assessing CRISPR-edited mammalian cell lines using a single-step 3-color Crystal Digital PCR™ assay
The generation and usage of homozygous fluorescent knock-in mammalian cell lines using sequence-specific Cas-endonucleases has become a commonly used molecular biology procedure.1-8 Nevertheless, to identify the desired genetic modification and reject clones with off-target events, several hundred monoclonal colonies still need to be screened. The maintenance and screening of high numbers of clones is a laborious, time-consuming, and error-prone process. To facilitate colony screening, we implemented a single-step 3-color assay using Crystal Digital PCR™ to assess on-target copy number and detect off-target events simultaneously. Because digital PCR (dPCR) is highly sensitive, it requires low amounts of biological samples. Thus, the screening assay was integrated early in the genome engineering pipeline. In addition, because the single-step 3-color Crystal Digital PCR™ assay is achieved in a single assay in about three hours, the culture of incorrectly edited clones can be discontinued on the same day and early in the pipeline
Single-step 3-color digital PCR assay design
- Total tag integration design | The “total-mEGFP” assay detects the presence of the mEGFP sequence in the genome. PCR primers and a green HEX fluorescent probe anneal to the mEGFP sequence and are designed to detect only the mEGFP transgene.
- On-target integration design | The “on-target-tag” assay detects the junction between the last exon of the endogenous NUP93 editing site and the 5’- sequence of the integrated mEGFP transgene (tag). PCR primers and a blue FAM fluorescent probe are designed to amplify and detect the “on-target tag” for the determination of scarfree 5’ integration.
- Reference design | The “reference” assay detects a sequence roughly 8-10 kb downstream of the target site, on the same chromosome. PCR primers and a red Cy®5 fluorescent probe anneal to the reference sequence and are designed to detect only the “reference” product, which is comparable in size to the “on target-tag” and “total-mEGFP” amplicons.
- Computation of total mEGFP copies | The number of integrated mEGFP copies was obtained by normalizing the copies (cp)/μL of the “total mEGFP” assay with the cp/μL of the “reference” assay.
- Computation of on-target mEGFP copies | The number of on-target integrated mEGFP copies was obtained by normalizing the cp/μL of the “on target tag” assay with the cp/μL of the “reference” assay.
Figure 1 | Single-step 3-color dPCR assay validation using CRISPR-edited U-2 OS Nup93-mEGFP clones.
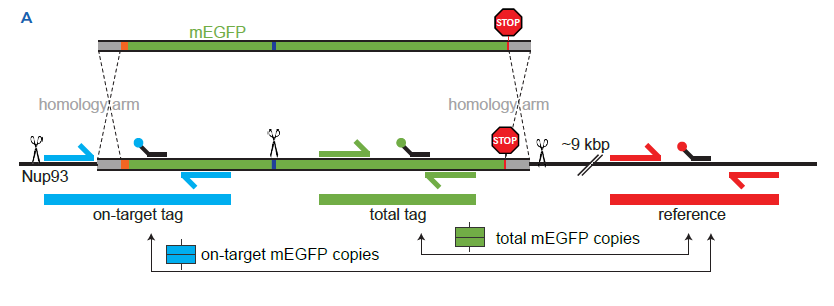
1A Donor design (top) | A PCR product, amplified from a synthetic dsDNA template, is used as donor. It includes a short sequence (~50 bp homology arm) homologous to the 5’-region of NUP93 (in grey), a linker (~5-10 amino acids, orange square), a STOP codon and a short sequence (50bp homology arm) homologous to the 3’-UTR of NUP93 (in grey). Notably, the mEGFP sequence contains a silent mutation to generate a restriction to have independent templates during digital PCR (deep blue bar). A STOP codon and a short sequence (50bp homology arm) homologous to the 3’-UTR of NUP93 (in grey). Single-step 3-color Crystal Digital PCR™ assay design (bottom) | Dashed line indicates homology-directed repair (HDR) after the targeted spCas9-mediated double-strand break (DSB). Primer positions are shown to generate products corresponding to “on-target tag,” “total mEGFP,” and “reference” assays. Arrows point to the ratios of products representing total mEGFP copies and on-target mEGFP copies.
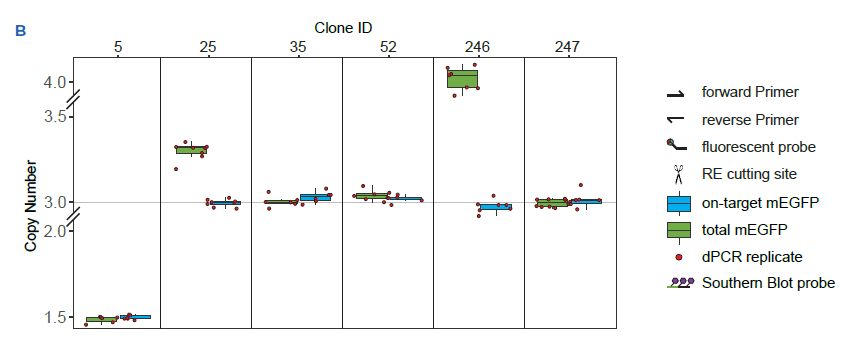
1B Crystal Digital PCR™ measurements of six representative clones | Boxes represent on-target (blue) and total mEGFP (green) copy numbers. Every red dot represents an individual dPCR measurement (technical replicates). Allelicity of the NUP93 gene in U-2 OS cells is three. Clones (clone ID 35, 52 and 347) scoring at 3 on-target and at 3 total mEGFP copies are homozygous for NUP93-mEGFP. A total number of mEGFP copies higher than 3 suggests extra integrations (clone ID 25 and 246). Estimates lower than 3 indicate either fewer tag integrations (monoallelic or biallelic clones) or potential genomic rearrangements (clone ID 5).
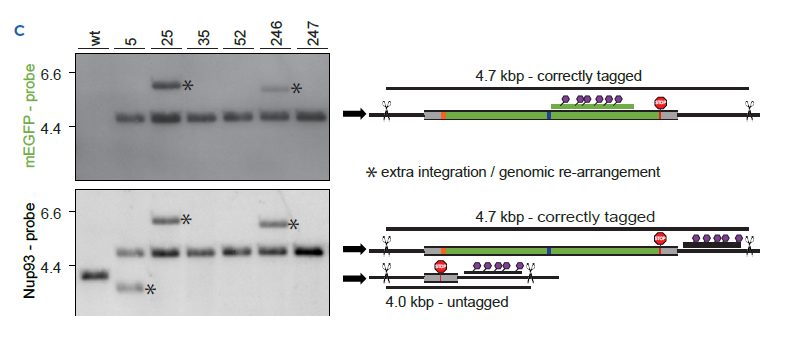
1C Southern Blot performed with the same clones as in B | The band at ~4.7kb indicates a correctly tagged NUP93-mEGFP allele detected with a mEGFP specific probe. Any additional band detected using the mEGFP-specific probe indicates an extra integration of the tag in the genome. The Nup93-specific probe reveals the correctly tagged NUP93-mEGFP alleles at ~4.7 kb and the untagged alleles at ~4.0 kb. Bands marked with an asterisk correspond to unexpected, extra integrations or major genomic re-arrangements.
Figure 2 | Representative 2D dot plots from Crystal Miner software
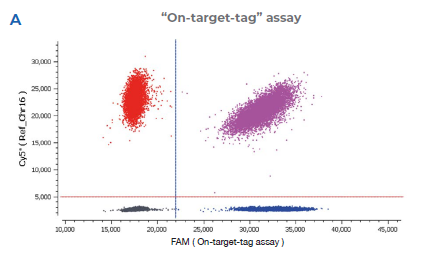
2A “On-target-tag” assay detected by the FAM probe (On-target-tag assay) that binds to the junction between the last exon of the endogenous NUP93 editing site and 5’ sequence of the integrated mEGFP transgene.
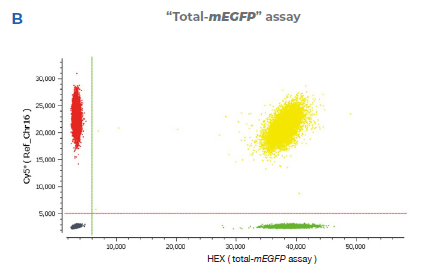
2B “Total-mEGFP” assay detected by the HEX probe (total-mEGFP assay) that binds to the mEGFP sequence and is designed to detect
only the mEGFP transgene. The reference assay is detected by a Cy®5 probe and binds to a sequence downstream of the target site on
the same chromosome (Chr16).
- Conclusion about the CRISPR-driven integration event (homozygous, monoallelic, biallelic, off-targets integration, error-prone integration) | If the ratio of the scores retrieved from the two assays (total mEGFP and ontarget- tag assay) is not equal to 1, then an adverse editing event occurred (such as off-target integration, mis-editing, or incomplete editing).
- An in-house developed “R” notebook allows automated copy number assessment of clones and is available at https://github.com/beatrizserrano/CRISPR_updated_protocol.
- The internal probes of the three assays were labeled with FAM, HEX or Cy®5.
Application Note Highlights
- Crystal Digital PCR™, provides a robust and quantitative tool to assess on-target copy number and detect off-target events in a single 3-color assay.
- Crystal Digital PCR™ allows robust copy number assessment of large insertions such as mEGFP.
- Crystal Digital PCR™ uses small amounts of biological material and can be introduced early in the CRISPR screening pipeline, thus saving time.
Notes
- The forward oligo of the “on-target-tag” assay should anneal outside of the homology arm of the CRISPR donor template
- Short homology arms should be designed (≤50bp) to optimize Crystal Digital PCR™ performance and accuracy
- All Crystal Digital PCR™ templates must be separated molecules. This can be achieved by restriction digestion of the genomic DNA. If a specific restriction site is not available within the donor template, a silent mutation can be generated to obtain an extra cutting site, as for mEGFP.
- The “reference” assay should be designed on the same chromosome as the on-target assay. Although unlikely to occur, an off-target event happening within the reference locus would lead to incorrect copy number estimates.
- A positive control for the “on-target-tag” and the “total- mEGFP” assays can be performed using wildtype genomic DNA – that is, unmodified – plus a donor plasmid carrying the desired genomic target region.
- See also the technical note: Guidelines for 3-color multiplex assay design for optimized performance with Crystal Digital PCR™ https://www.stillatechnologies.com/digital-pcr/naica-system-analysis/guidelines-for-3-color-multiplex-assay-design-for-optimized-performance-with-crystal-digital-pcr/
Results
We have developed a single-step 3-color Crystal Digital PCR™ assay and assessed its predictive power by comparing it to a conventional Southern blot. There are three NUP93 alleles in U-2 OS cells. This quantitative assay showed that clones 35, 52 and 247 scored 3 on-target and total mEGFP copies, meaning that all three NUP93 alleles were successfully tagged (i.e. homozygous clones), with no detectable off-target integrations. The Southern blot confirmed this conclusion. Clone 5 shows an equal number of mEGFP copies for both assays, though without matching the expected number of NUP93 loci. The copy number assessment may indicate a single, correct editing event accompanied by a major genomic re-arrangement, as suggested by Southern Blot analysis. Clone 25 and 246 show a high number of total mEGFP integrations vs on-target integrations, indicating potential off-target editing or incomplete HDR. Southern Blot displays extra bands detected with the mEGFP and NUP93 probes, suggesting genome rearrangements. Results obtained by 3-color Crystal Digital PCR™, a straightforward one-step detection assay, were perfectly concordant with the more labor-intensive southern blot technique.
Bibliography
- Kueblbeck, M., Callegari, A., Serrano-Solano, B. & Ellenberg, J. Rapid generation of homozygous fluorescent knock-in human cells using CRISPR/Cas9 genome editing and validation by automated imaging and digital PCR screening AUTHOR CONTRIBUTIONS. bioRxiv 2021.06.23.449557 (2021) doi:10.1101/2021.06.23.449557.
- Cai, Y. et al. Experimental and computational framework for a dynamic protein atlas of human cell division. (2018) doi:10.1038/s41586- 018-0518-z.
- Walther, N. et al. A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J. Cell Biol. 217, 2309–2328 (2018)
- Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017).
- Cattoglio, C. et al. Determining cellular CTCF and cohesin abundances to constrain 3D genome models. Elife 8, (2019).
- Okamoto, S., Amaishi, Y., Maki, I., Enoki, T. & Mineno, J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs OPEN. doi:10.1038/s41598-019-41121-4.
- Liang, X., Potter, J., Kumar, S., Ravinder, N. & Chesnut, J. D. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J. Biotechnol. 241, 136–146 (2017).
- Roth, T. L. et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018).
To learn more about digital PCR, please visit Stilla Technologies’ Learning Center.
Check out our joint webinar: https://www.youtube.com/watch?v=ErEr5xyCWek
moritz.kueblbeck@embl.de | andrea.x.callegari@gsk.com | beatriz.serrano-solano@embl.de