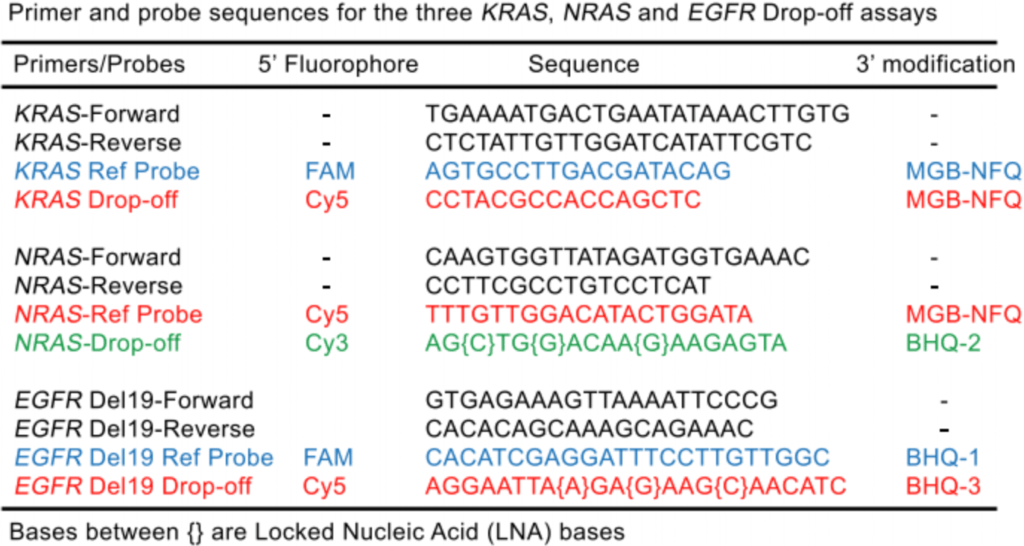
Drop-off Crystal Digital PCR™ for NRAS, KRAS, & EGFR mutations
How to design and quantify using a drop-off assay
A major advantage of a drop-off digital PCR is the single-assay detection of multiple proximal genetic lesions (including deletions, insertions and nucleotide substitutions) within a short genomic interval. The most simplified version of a drop-off assay includes two TaqMan™ probes targeting the same amplicon: a Drop-off Probe that spans the mutation hotspot but is uniquely complementary to the wild-type sequence, and a Reference Probe that hybridizes adjacent to the mutation site and is complementary to both the mutant and the wild-type alleles (Figure 1A). In the presence of a wild-type allele, both the drop-off and reference probes will hybridize with their target, leading to a double positive signal (Figure 1B, turquoise population). By contrast, in the presence of a mutant allele, even a single nucleotide mutation is enough to destabilize the hybridization of the drop-off probe so that only the reference probe anneals to its target leading to a simple positive signal (Figure 1B, green population).
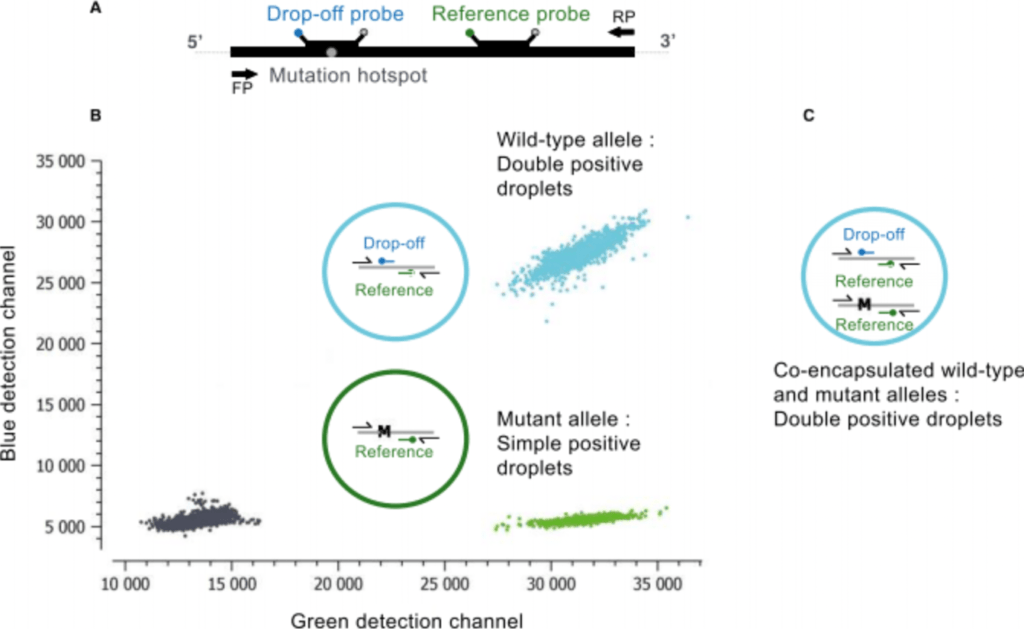
Figure 1: A. Schematic of the hybridization positions of the Drop-off and Reference probes and the primer pair (arrows) on the amplified target (FP=forward primer; RP=reverse primer). B. Crystal Miner 2D dot-plot of a drop-off assay showing clusters of fluorescent droplets derived from double positive wild-type alleles (turquoise) and simple positive mutant (M) alleles (green). Double negative droplets are shown in black (C). It is important to note that during droplet generation, a fraction of mutant alleles and wild-type alleles could be randomly co-encapsulated within the same droplet rendering the droplet double positive wild-type and mutant (turquoise). The wild-type concentration can be directly derived from the proportion of droplets that are double positive (regardless of whether they are positive or negative in the 3rd color channel). The mutant concentration can be derived from the proportion of droplets that contain mutant alleles (regardless of whether they are positive or negative in the 3rd channel), but this proportion can only be determined by discarding the double positive droplets. Indeed, if the double positive droplets are counted as mutant negative droplets, it would underestimate the mutant allele frequency. For more details on how to quantify a Drop-off assay, please see our Technical Note on Drop-off Quantification at www.stillatechnologies.com/technical-notes/ and Whale, AS, et al., Biomol Detect Quantif. 2016 27; 10: 15-23.
Drop-off assays detect KRAS, NRAS and EGFR hotspot mutations
In clinical settings, a set of predictive genetic markers are routinely monitored to track therapy efficacy. For example, in non-small cell lung cancer the presence of deletions in the epidermal growth factor (EGFR) exon 19 confers sensitivity to first generation tyrosine kinase inhibitors. Moreover, in colorectal carcinoma, KRAS and NRAS proto-oncogene mutations are strong indicators of resistance to anti-EGFR antibodies. Using drop-off assays and the 3-color multiplexing capacity of the naica® system, three internally controlled drop-off digital PCR assays were designed to detect the most prevalent KRAS exon 12, NRAS exon 3 and EGFR exon 19 sequence alterations (Figures 2 and 3). The addition of an internal control (IC) allows the straightforward evaluation of assay robustness and the identification of PCR inhibition, which can occur due to sample impurity.
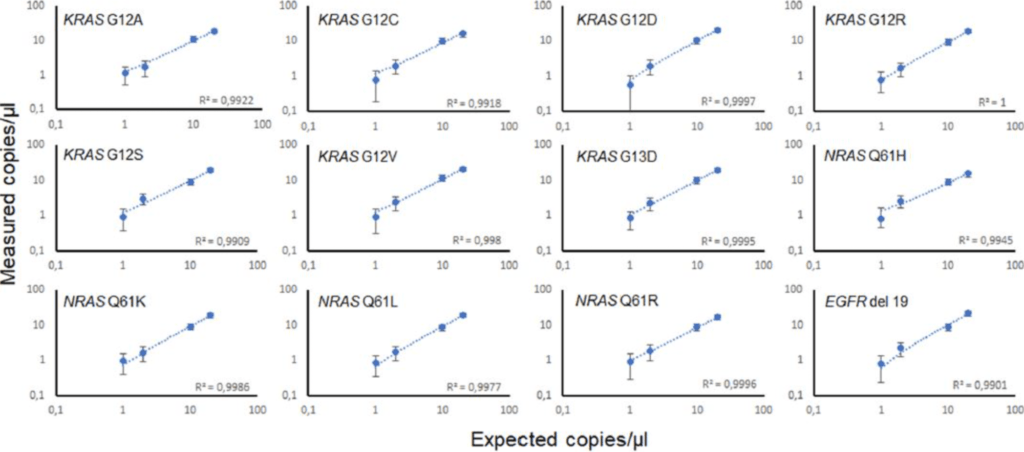
Figure 2: Reliability of drop-off assays for the detection of the seven and four most prevalent KRAS exon 12/13 and NRAS exon 3 mutations, respectively, and EGFR exon 19 deletions. The mutations were detected with a 95% confidence level in serial dilutions ranging from 5% to 0.25 % of mutant DNA at final concentrations down to 1 copy/µl in a 25µl PCR mixture. All assays were performed in a reaction background of 104 copies of wild-type DNA and 400 copies of the internal positive control DNA (ΦX174 bacteriophage). N=3 replicates for each dilution point. The displayed confidence intervals are the means of the theoretical confidence intervals accounting for sampling and partitioning error at a 95% confidence level.
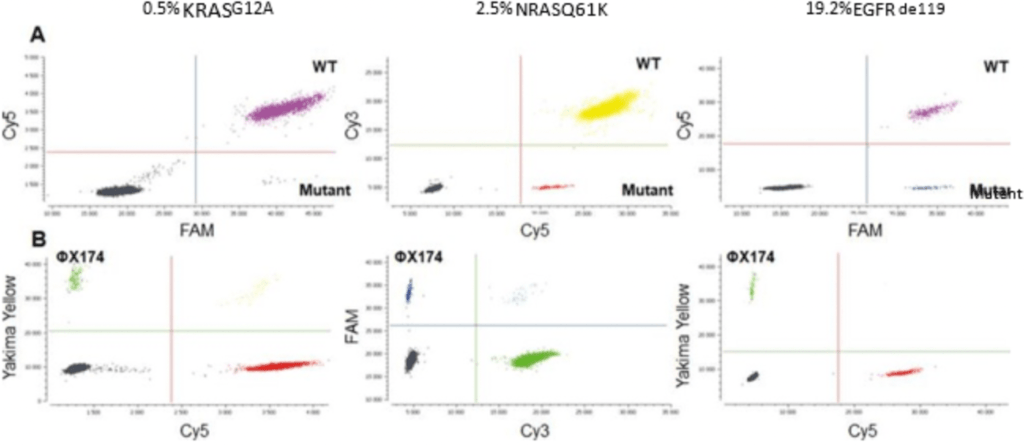
Figure 3: A. Crystal Miner-generated 2D dot plots of the KRAS, NRAS and EGFR drop-off digital PCR assays in triplex experiments. Commercial DNA (KRAS and NRAS), as well as DNA derived from frozen tumor samples (EGFR) were used. WT: Wild-Type. B. Dotplots displaying the signal obtained in the third-color channel for each assay using the ΦX174 DNA internal positive control detection assay.
As multiple mutations are known to occur in KRAS and NRAS hotspots, and several deletions/insertions of varying lengths have been described in EGFR exon 19, the use of drop-off assays allows rapid and cost-efficient simultaneous screening of a variety of clinically relevant genetic alterations using a limited number of probes. If desired, once a drop-off assay has identified a sample as mutant, subsequent assays using sequence-specific probes can be employed to determine the exact mutant alleles existing within the DNA sample.
Application Note Highlights
- Drop-off digital PCR assays enable the simultaneous detection of multiple mutations occurring at genomic hotspots.
- Drop-off assays allow rapid and cost-efficient screening of a variety of genetic alterations using a limited number of probes.
- Using Crystal digital PCR™ we designed and validated three internally controlled drop-off assays for the detection of seven KRAS mutations, four NRAS mutations and a range of EGFR exon 19 deletions/insertions commonly monitored in clinical practice.