A streamlined workflow for liquid biopsy sample extraction and highplex Crystal Digital PCR™ analysis using the Maxwell® RSC system and the 6-color naica® system
Introduction
Liquid biopsies, such as blood samples, can harbor a wealth of genetic information from both healthy and unhealthy cells to inform disease diagnosis and treatment. Rigorously qualified pre-analytical protocols are vital to ensure the performance of downstream liquid biopsy workflows and thus high-quality results. Circulating cell free DNA (cfDNA), extracted from liquid biopsy samples, are an established sample type for characterizing oncology targets. Nevertheless, cfDNA measurements require a highly sensitive and reliable detection technology to quantify, often low-level, genetic aberrations within a complex background of wild-type sequences.
This technical note details a flexible method for automated plasma sample extraction that seamlessly integrates into a straightforward and sensitive digital PCR genetic analysis workflow. Combining the Maxwell® RSC 48 instrument for automated cfDNA extraction with Crystal Digital PCR™ on the naica® system allows ultrasensitive high-plex detection from liquid biopsy samples. By bridging the two technologies, 32 common and rare somatic EGFR mutations in exons 18, 19, 20, and 21, representing more than 90% of EGFR mutations described in non-small-cell lung carcinoma (NSCLC) are sensitively and precisely detected. The combination of the two technologies also allowed for the detection of a set of PIK3CA mutations from cfDNA samples.
The combined workflow of automated plasma sample extraction using the Maxwell® RSC 48 and the high-plex and sensitive target detection with Crystal Digital PCR™ thus represents a streamlined full solution from sample-to-answer that can benefit cancer researchers across the biomarker testing landscape.
Materials and methods
Plasma-like samples (SensID reference material ref SID- 000002, SID-000016, SID-000089) and human K2EDTA plasma samples, collected from healthy donors, were extracted with the Promega Maxwell® RSC ccfDNA LV Plasma Kit (Promega, ref AS1840). Before extraction, all samples were spiked with a known quantity of an exogenous extraction control DNA (EC), and after extraction, aliquots of the human plasma samples were spiked with known amounts of synthetic mutant DNA. For comparison to the automated sample extraction methodology, plasma samples were also manually extracted with the QIAamp circulating nucleic acid kit (QIAGEN, ref 55114) according to supplier recommendations.
The extracted cfDNA samples were then analyzed by Crystal Digital PCR™ on the 6-color naica® system with two independent multiplexed cancer detection assays, a 6-plex and a 33-plex.
Stilla Technologies defines plex as the number of targets detected in a Crystal Digital PCR™ assay.
A custom 6-plex Rectal Cancer assay detecting six targets: PIK3CA wild-type, four PIK3CA mutations (p.E542K, p. E545K, p.H1047L, p. H1047R) and the EC.
The EGFR 6-color Crystal Digital PCR™ kit (Stilla Technologies®, Ref R30006), an off-the-shelf 33-plex digital PCR kit detecting EGFR wild-type and the 32 most common non-small cell lung cancer EGFR mutations1.
Maxwell® RSC 48 Instrument
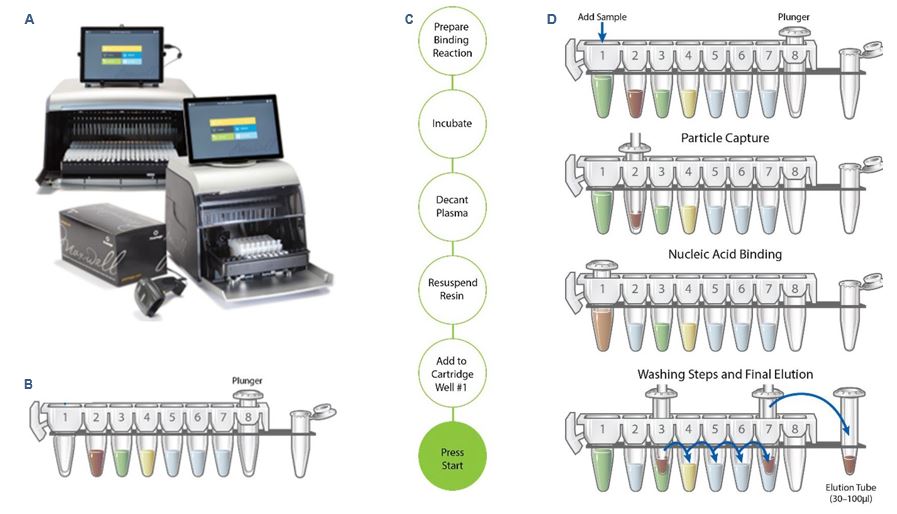
The Maxwell® RSC 48 Instrument (Figure 1A) is a compact, automated nucleic acid purification platform that processes up to 48 samples simultaneously. Using Maxwell® cartridges, prefilled with purification reagents and paramagnetic particles (Figure 1B), the Maxwell® RSC 48 Instrument enables consistent high-throughput purification of DNA or RNA from a variety of sample types. The Maxwell® RSC 48 Instrument’s intuitive graphical interface makes the instrument easy to use. The integrated vision system with its large LED indicator reduces the potential for user error by detecting proper cartridge placement. This informs the user of any issues before the run starts. An integrated bar code reader makes it easy to track sample information along the extraction workflow.
Because the Maxwell® RSC instruments are magnetic particle movers, not liquid handlers, they offer advantages over other automated nucleic acid extraction systems. There is minimal risk of cross-contamination because no liquid handling or splashing occurs during sample processing. With no clogs and fewer breakdowns, there are fewer disruptions to the nucleic acid extraction workflow. High-quality nucleic acid purification, with minimal steps and hands-on time, can be obtained with a wide range of available extraction kits, including the Large Volume (LV) ccfDNA plasma extraction kit used for this study (Figure 1C and 1D).
Crystal Digital PCR™ on the 6-color naica® system

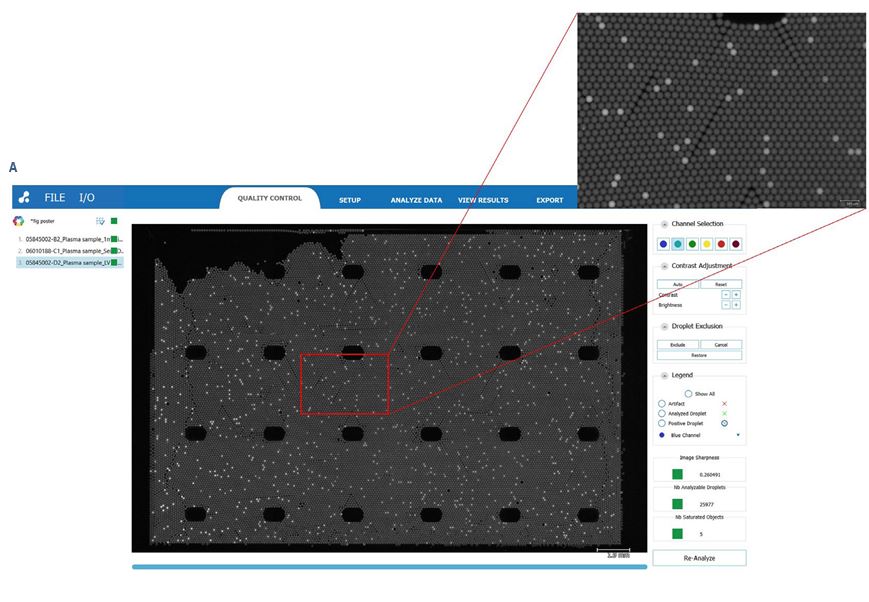
The 6-color naica® system Crystal Digital PCR™ workflow (Fig- ure 2) enables high multiplexing capacity in a single reaction, saving both time and precious samples, and provides ultra-sensitivity and increased low-level detection of multiple reactions in parallel. By automatically partitioning sample reactions into a 2D array of tens of thousands of droplets, Crystal Digital PCR™ technology can be used for absolute nucleic acid quantification in a wide range of applications. For example, Crystal Digital PCR™ can be used for oncological analysis applications, including copy number variation, mutation detection, rare event detection, and therapeutic monitoring. The accompanying Crystal Miner software measures the concentrations of targeted nucleic acids, providing automatic identification of positive and negative Droplet Crystals for the selected fluorescence channels. With intuitive visuals for image analysis, users can easily explore their multiplex data and directly inspect the Droplet Crystals in different fluorophore channels. This visual inspection allows unparalleled peace of mind for quality control.
Results
Integration of the Maxwell® RSC LV kit and the 6-color naica® system into a compatible sample-to-answer workflow
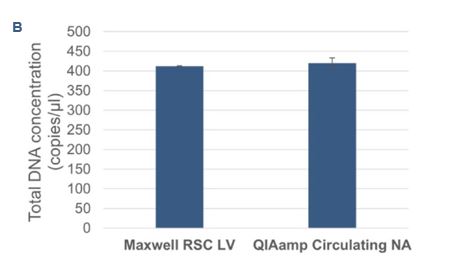
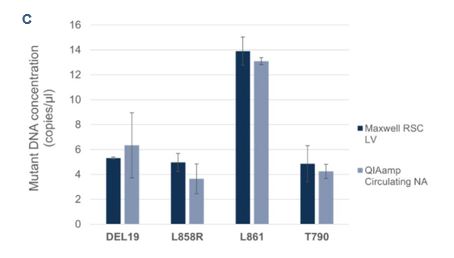
To evaluate the Droplet Crystal stability and compare the sample concentrations of cfDNA using two different extraction procedures, cfDNA from plasma-like samples were extracted automatically with the Maxwell® RSC LV kit on the Maxwell® RSC 48 system (Promega) and manually with the QIAamp circulating nucleic acid kit (Qiagen). The samples were processed by Crystal Digital PCR™ on the 6-color naica® system using the EGFR 6-color Crystal Digital PCR™ kit. All samples showed full compatibility for Droplet Crystal stability (Figure 3A). Furthermore, a comparison of the total DNA concentration (Figure 3B) and mutant DNA concentrations (Figure 3C) revealed highly similar results across the two extraction procedures.
Mutation detection in plasma and plasma-like samples
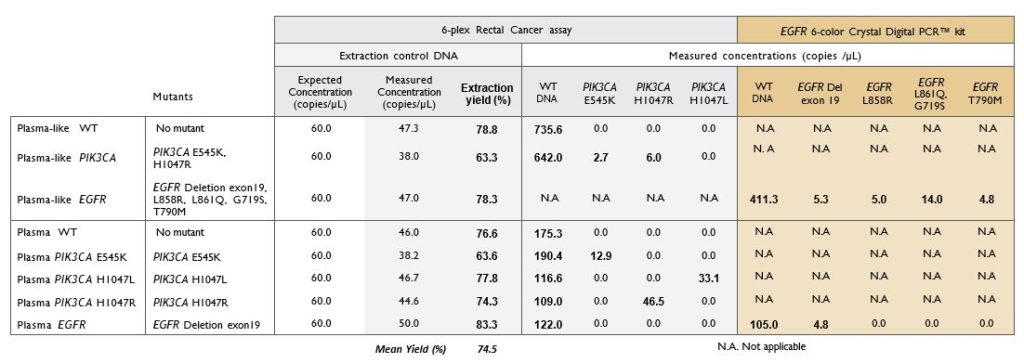
Extracted cfDNA from both plasma and plasma-like samples were analyzed by two multiplexed detection assays customized for Crystal Digital PCR™ workflow: a custom designed 6-plex Rectal Cancer assay and the commercially available EGFR 6-color Crystal Digital PCR™ kit. Quantification of the EC allowed to calculate the extraction yield for each sample. The mean yield obtained for eight samples (three plasma-like and five plasma) was 74.5%. All mutations known to be present in the samples were detected (Table 1).
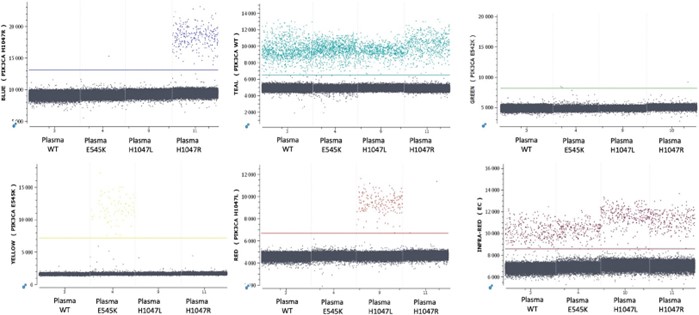
Example 1D-dot plots generated by the Crystal Miner software resulting from the detection by the 6-plex Rectal Cancer assay are shown in Figure 4.
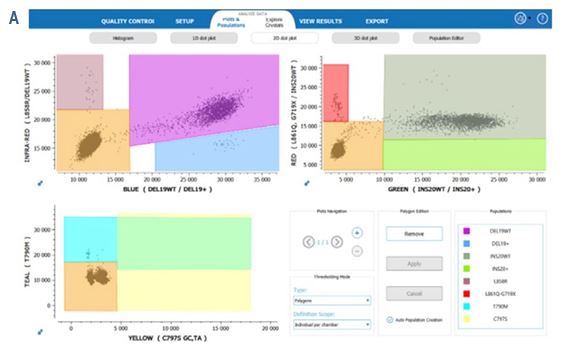
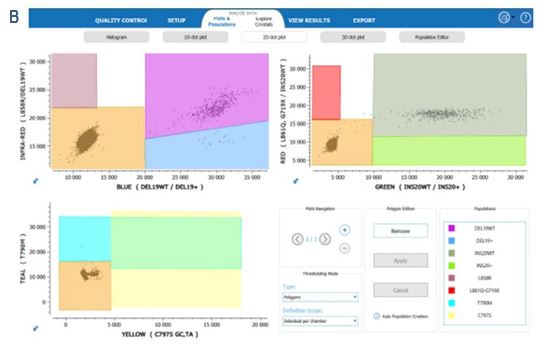
Furthermore, 2D-dot plots resulting from the detection by the EGFR 6-color Crystal Digital PCR™ kit show clear separability between the positive and negative clusters. (Figure 5).
CONCLUSIONS
This study demonstrates compatibility between the automated Maxwell® RSC cfDNA extraction system and high-plex detection from liquid biopsy samples by Crystal Digital PCR™ on the 6-color naica® system. This full solution workflow enables largely automated highly sensitive detection and quantification of the main somatic EGFR mutations described in NSCLC and a set of PIK3CA mutations in breast and rectal cancers. Both technologies produce results quickly with minimal hands-on-time, enabling a complete sample-to-result workflow in less than a day. This proof-of-concept workflow creates the foundation for the further development of streamlined sample-to-answer protocols for high multiplex assays that will benefit cancer researchers across the biomarker testing landscape.
HIGHLIGHTS
- The Maxwell® RSC cfDNA extraction system and Crystal Digital PCR™ on the 6-color naica® system for a seamless, largely automated, highly sensitive detection and quantification end-to-end workflow.
- Demonstrated detection and quantification of the main somatic EGFR mutations described in NSCLC and a set of PIK3CA mutations described in breast and rectal cancers.
- Minimal hands-on-time, enabling a complete full solution sample-to-result workflow in less than a day.
For further information on Maxwell Extraction systems and technologies, contact Maxwell-dPCR@Promega.com
Reference:
1 Harrison PT, Vyse S, Huang PH, Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer, Seminars in Cancer Biology (2019), doi: https://doi.org/10.1016/j.semcancer.2019.09.015
Learn more about what the Nio®+ instrument and Crystal Digital PCR™ can do for your research

