Detection of hypermethylated circulating tumor DNA by Crystal Digital PCR™
In collaboration with:
- Dr. Valérie Taly, Université de Paris, Centre de Recherche des Cordeliers, Centre Nationale de la Recherche Scientifique (CNRS)
- Geoffroy Poulet, Université de Paris, Eurofins Biomnis
- Pr. Pierre Laurent-Puig, Université de Paris, Centre de Recherche des Cordeliers, APHP Hôpital Européen Georges-Pompidou

Hypermethylated WIF1 and NPY as biomarkers in colorectal cancer
Changes in the DNA methylation status of gene regulatory regions constitute emerging biomarkers for a variety of cancers. This epigenetic alteration is biologically stable and present in circulating tumor DNA, making it suitable for early detection and noninvasive dynamic monitoring of tumor burden. In colorectal cancer (CRC), hypermethylation of WNT inhibitory factor 1 (WIF1) and neuropeptide T (NPY) was found in 80% and 44.7% of metastatic and stage II/III patients, respectively [1]. To evaluate if hypermethylated WIF1 and NPY can be used as a universal colorectal cancer marker and a surrogate to tumor sequence-specific mutations, a method combining bisulfite conversion (Figure 1.A) of unmethylated cytosine to uracil and Crystal Digital PCR was developed. A 3-color digital PCR assay targeting bisulfite converted hypermethylated promoter regions of WIF1 and NPY and unmethylated Albumin gene ALB as a reference was optimized and tested on DNA extracted from the plasma of healthy individuals and CRC patients (Figure 1.B).
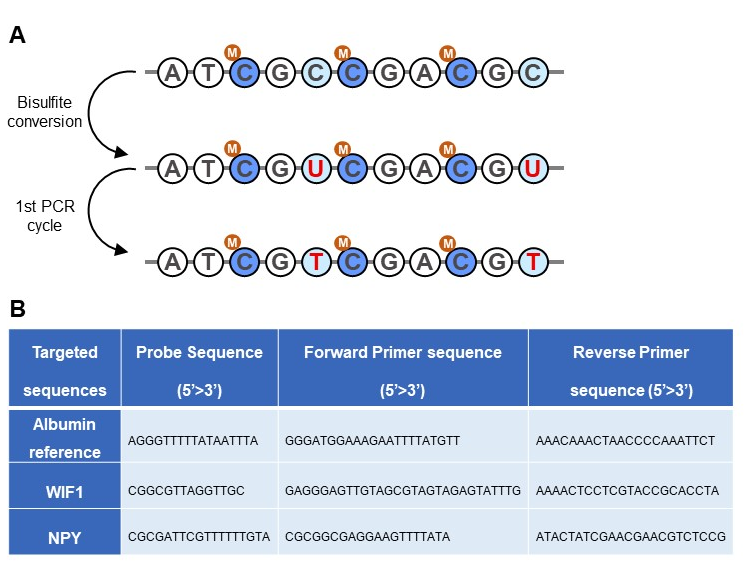
Figure 1: A. Bisulfite conversion of unmethylated cytosine (C) residues leads to their deamination to uracil (U) while methylated cytosine (M) residues (present in CpG islands) are protected from bisulfite conversion. PCR detection recognizes the uracil products of this conversion as thymine (T), thus distinguishing them from unconverted (methylated) cytosine. B. Sequences of the primers and fluorescent-labeled probes used for the 3-color Crystal Digital PCR assay [1]. Between 17 and 24 CpG islands displaying a similar methylation profile are targeted by the WIF1 and NPY amplicons.
A 3-color Crystal Digital PCR assay detects hypermethylated WIF1 and NPY biomarkers
The limit of blank (LOB) with a 95% confidence level of the triplex Crystal Digital PCR assay, defined as 3 and 12 false positive droplets for NPY and WIF1, respectively, was determined on 31 sample replicates containing unmethylated DNA ranging from 40 to 400 cp/µl (corresponding to 1000 to 10,000 copies per 25 µl reaction). To evaluate the sensitivity and the linearity of the assay, serial dilutions of chemically methylated DNA ranging from 20 to 0.2 cp/µl in a background of 400 cp/µl of unmethylated DNA (representing 5 to 0.05 % of the mutant allele fraction) were assayed in triplicate. For NPY and WIF1, a total of 0.2 cp/µl and 1 cp/µl (5 and 25 copies of methylated DNA copies per 25µl reaction) were reliably detected, respectively, corresponding to a mutant allele fraction of 0.05% and 0.25% (Figure 2).
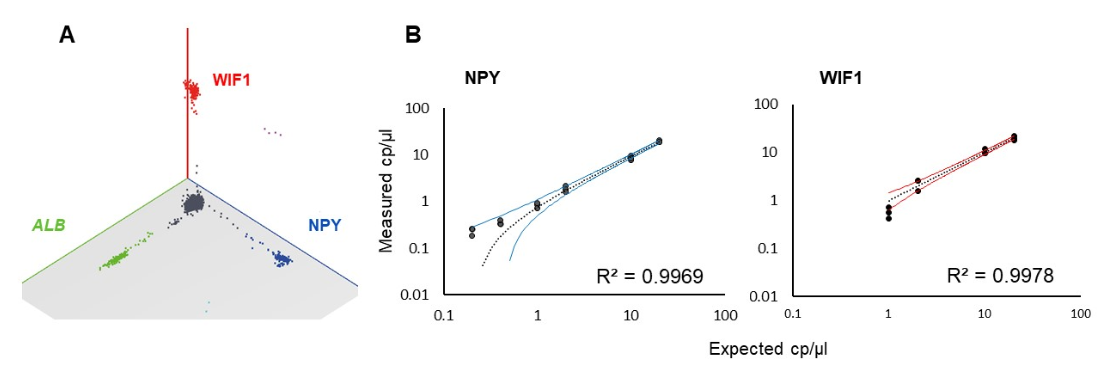
Figure 2: A. Crystal Miner 3D dot-plot of the 3-color Crystal Digital PCR assay for the detection of hypermethylated WIF1, NPY and ALB on chemically methylated DNA. B. Standard curves obtained by measuring serial dilutions of methylated DNA in a background of unmethylated DNA. To determine whether the observed mutant quantity was significantly above the LOB, a Bayesian approach based on false positive probability was used. For each result, the number of positive partitions was corrected by deducting eventual false-positive partitions weighted by their probability distribution. A sample was considered as negative when the lower bound of the corrected 95% confidence interval included zero, in which case it is not represented on the graph. For more information, please see here.
Detection of hypermethylated WIF1 and NPY in cancer patients and healthy individuals
A total of 10 and 5 DNA samples derived from plasma of stage III or IV CRC patients and healthy individuals, respectively, was tested using the 3-color Crystal Digital PCR assay. All plasma DNA samples from CRC patients scored positive for hypermethylated WIF1 and NPY, whereas hypermethylated WIF1 and NPY were not detected in healthy individuals. The fraction of hypermethylated plasma DNA was calculated by reporting the WIF1 and NPY concentration to the unmethylated ALB reference concentration. The hypermethylated WIF1 fraction in plasma DNA ranged from 8% to 93%, while that of hypermethylated NPY ranged from 0.1% to 78% (Figure 3). The lowest quantity of hypermethylated WIF1 and NPY detected in plasma samples was 5.1 and 1.2 copies per µl of bisulfited DNA eluate, respectively.
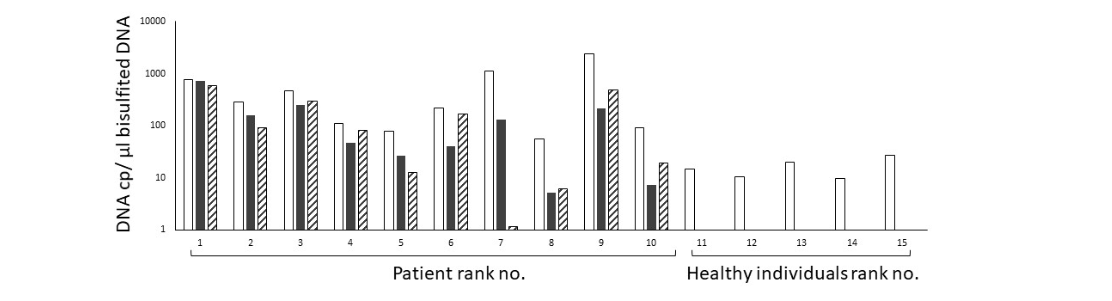
Figure 3: Concentration of ALB (white bars), hypermethylated WIF1 (black bars) and hypermethylated NPY (hashed bars) in plasma of CRC patients and healthy individuals. DNA concentration is expressed in copies per µl of bisulfited DNA eluate. Depending on the DNA concentration of the sample, 20ng or 20µl of DNA sample extracted from the patients was bisulfite-treated in a thermocycler at 98 °C for 10 minutes and 53 °C for 4 hours using the EZ DNA Methylation-Gold kit (Zymo Research) and eluted in 10µl final volume.
[1] Garrigou S, Perkins G, Garlan F, Normand C, Didelot A, Le Corre D, Peyvandi S, Mulot C, Niarra R, Aucouturier P, Chatellier G, Nizard P, Perez-Toralla K, Zonta E, Charpy C, Pujals A, Barau C, Bouché O, Emile JF, Pezet D, Bibeau F, Hutchison JB, Link DR, Zaanan A, Laurent-Puig P, Sobhani I, Taly V. A Study of Hypermethylated Circulating Tumor DNA as a Universal Colorectal Cancer Biomarker. Clin Chem. 2016 Aug;62(8):1129-39.
Application Note Highlights
- Bisulfite conversion followed by 3-color Crystal Digital PCR enables the reliable detection of down to 25 and 5 copies of hypermethylated WIF1 and NPY DNA, respectively, per 25µl reaction
- Crystal Digital PCR detection of hypermethylated WIF1 and NPY can be used as a universal colorectal cancer marker and a surrogate to tumor- specific mutations
- Hypermethylated WIF1 and NPY were detected in all 10 stage III/IV colorectal cancer patient plasma samples, while hypermethylated WIF1 and NPY were not detected in any of the 5 healthy individuals.
To learn more about digital PCR, please visit Stilla Technologies’ Learning Center