High multiplex, ultrasensitive EGFR detection using the EGFR 6-color Crystal Digital PCR™ kit
Ready-to-use | Intuitive | Rapid time to results | High multiplexing | High sensitivity | ctDNA compatible
Single assay detection of more than 90% of described EGFR mutations in NSCLC
A liquid biopsy is the sampling and analysis of a non-solid biological specimen, most often the case blood. Liquid biopsies are a minimally invasive and straightforward sampling approach that can overcome the heterogeneity of tumors, and thus represent a valuable source of circulating tumor DNA (ctDNA) for oncological biomarker analysis.
ctDNA measurements require a highly sensitive and reliable detection technology to quantify often low-level genetic aberrations within a high background of wild-type sequences. Digital PCR has emerged as a powerful technology for the next-generation analysis of liquid biopsies.
Stilla® Technologies’ 6-color naica® system is an ultrasensitive digital PCR technology capable of simultaneously and precisely quantifying high numbers of biomarkers in a single sample, simplifying the detection process, minimizing detection variability, and significantly reducing the hands-on-time to results.
Non small cell lung cancer (NSCLC) is a leading cause of cancer mortality worldwide. Epidermal growth factor receptor (EGFR) is frequently mutated and is a well known genetic aberration in NSCLC. The highly multiplexed, ready-to-use EGFR 6-color Crystal Digital PCR™ kit allows the detection from circulating free DNA (cfDNA) of more than 90% of EGFR mutations described in NSCLC, including 32 common, rare, activating, and resistant somatic EGFR mutations in exons 18, 19, 20 and 21 (Table 1).
The EGFR 6-color Crystal Digital PCR™ kit enables highly multiplexed cfDNA analysis on the naica® system
The EGFR 6-color Crystal Digital PCR™ kit uses TaqMan™ probe technology to detect eight, point mutations, as well as twenty-four Exon 19 deletion and Exon 20 insertion alterations using drop-off detection (Figure 1).
Table 1 | EGFR mutations detectable with the EGFR 6-color Crystal Digital PCR™ kit
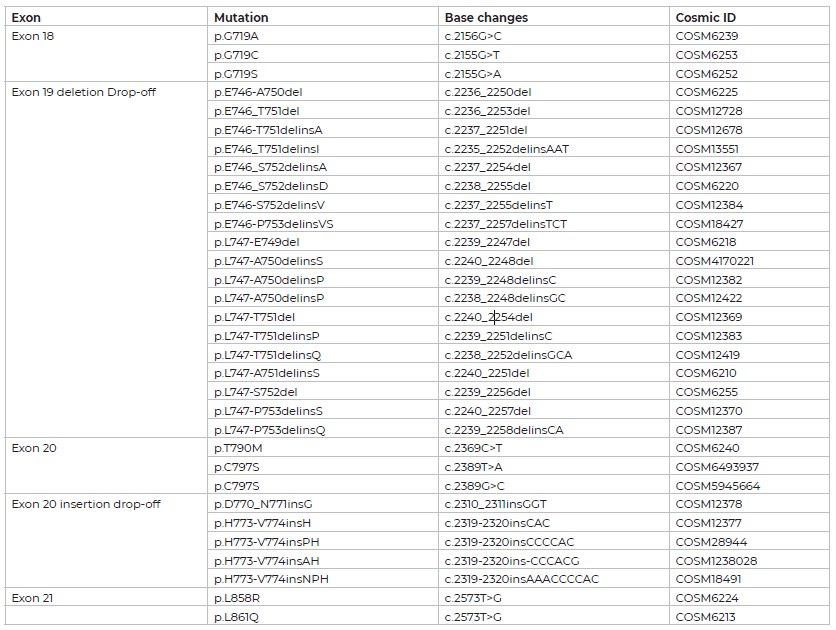
Figure 1 | EGFR 6-color Crystal Digital PCR™ kit detection design. TaqMan™ probe positions on each exon and fluorescent color code are noted.
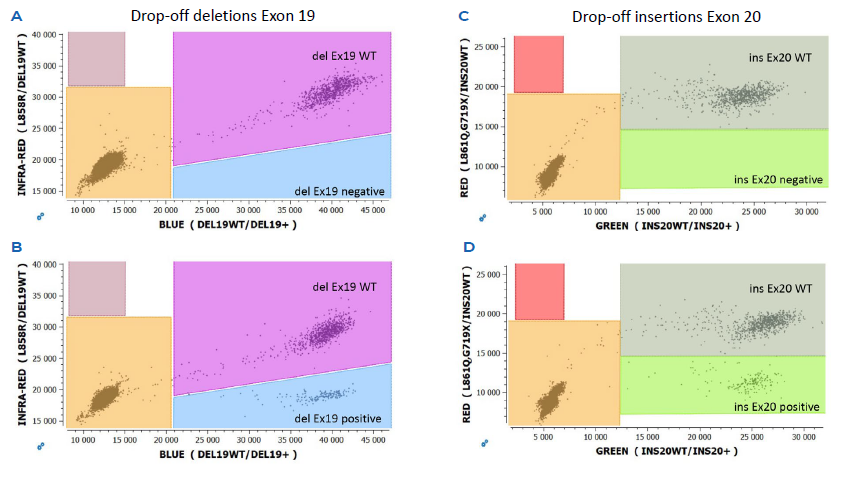
A major advantage of a drop-off digital PCR assay is the simplified detection of numerous proximal genetic lesions (including deletions, insertions and nucleotide substitutions) within a short genomic interval using minimal reagents. Indeed, a drop-off assay requires only two TaqMan™ probes targeting the same amplicon: a wild-type probe that spans the genetic lesion site but is uniquely complementary to the wildtype sequence, and a reference probe that hybridizes adjacent to the mutation site and is thus complementary to both the mutant and the wild-type alleles. In the presence of a wild-type allele, both the wild-type and reference probes will hybridize with their targets, leading to a double positive population (Figure 2A and 2C). Whereas in the presence of a mutant allele, only the reference probe anneals to its target leading to an additional simple positive population. (Figure 2B and 2D). The custom analysis template provided with the EGFR 6-color Crystal Digital PCR™ kit combined with Crystal Miner software allows the automated generation of 2D-plots and sample quantification.
Figure 2
A: Representative 2D-plots of wild-type (WT) deletion Exon 19 and
B: and deletion Exon 19 positive cfDNA samples
C: Representative 2D-plots of WT insertion Exon 20 and
D: and insertion Exon 20 positive cfDNA samples.
The x- and y-axis correspond to the fluorescence units of the indicated color channel.
The EGFR 6-color Crystal Digital PCR™ kit is optimized for ultrasensitive and highly specific detection on the naica® system.
The EGFR 6-color Crystal Digital PCR™ kit is compatible with the 6-color naica® system using highly sensitive Sapphire chips. To determine the sensitivity of detection of each EGFR target, the Limit of Blank (LoB) and the Limit of detection (LoD) were determined following the Clinical and Laboratory Standards Institute (CLSI) EP17-A2 standard (Protocols For Determination Of Limits Of Detection And Limits Of Quantitation; Approved Guideline). The LoB for EGFR mutation detection in the EGFR 6-color Crystal Digital PCR™ kit ranges from 0.06 to 0.17 copies per μL (cp/μL), depending on the target, whereas the LoD ranges from 0.30 to 0.46 cp/μL, depending on the target (Table 2).
Table 2 | LoB and LoD of the EGFR 6-color Crystal
Digital PCR™ kit determined following the CLSI EP17-A2 standard (Sapphire chip well concentrations in cp/μL).
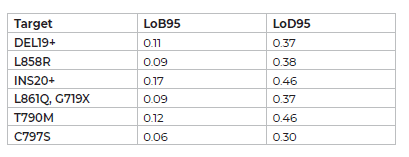
To evaluate the sensitivity of the EGFR 6-color Crystal Digital PCR™ kit, a linear titration was performed targeting EGFR exon 19 deletions, EGFR exon 20 insertions, EGFR L858R, EGFR G719A, EGFR L861Q, EGFR T790M, EGFR C797S (GC) and EGFR C797S (TA) mutations from 64 cp/μL down to 0.25 cp/μL (Figure 3) in a background of 120 cp/μl of wild-type EGFR DNA (equivalent to 3,000 copies per 25μl Sapphire chip reaction). The EGFR 6-color Crystal Digital PCR™ kit displayed ultrasensitive and reliable detection for all mutation.
Figure 3
Titrations of theoretical concentrations of 64cp/μl, 32 cp/μl, 16 cp/μl, 8 cp/μl, 4 cp/μl, 2 cp/μl, 1 cp/μl, 0.5 cp/μl and 0.25 cp/μl of EGFR exon 19 deletion, EGFR exon 20 insertion, EGFR L858R, EGFR G719A, EGFR L861Q, EGFR T790M, EGFR C797S (GC) and EGFR C797S (TA) DNAs, in a background of 120 cp/μl of wild-type EGFR (3,000 copies per 25μl reaction). Reactions were performed using synthetic Ultramer™ mutant DNA oligonucleotides templates ranging in size from 97 to 124 nucleotides, and wild-type human genomic DNA. Shown are the averages of a minimum of triplicate measurements with standard errors for each target.
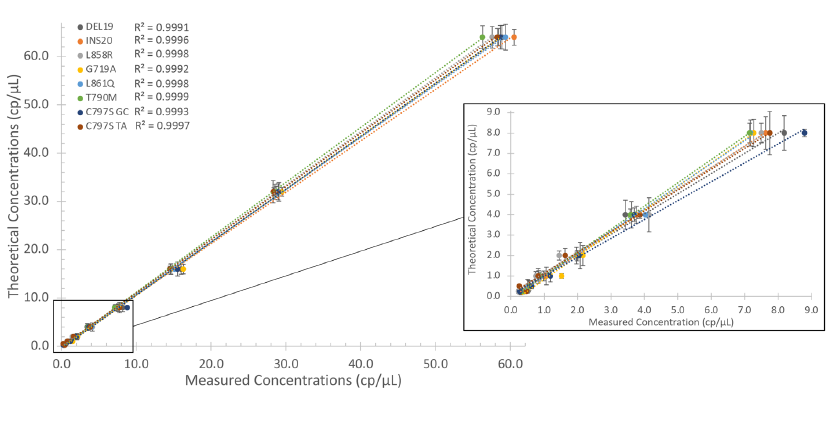
The EGFR 6-color Crystal Digital PCR™ kit showed robust inter-run repeatability, with concentration RSDs (relative standard deviations) ranging from 4.5 to 8.6% (N = 12), depending on the target (Figure 4). In addition, the EGFR 6-color Crystal Digital PCR™ kit showed high specificity, with no false positives obtained (Table 3).
Figure 4
Inter-run repeatability of the EGFR 6-color Crystal Digital PCR™ kit. The inter-run repeatability was measured by comparing the results obtained on one 1 positive control sample chamber in 12 different runs (N = 12) with the EGFR 6-color Crystal Digital PCR™ kit
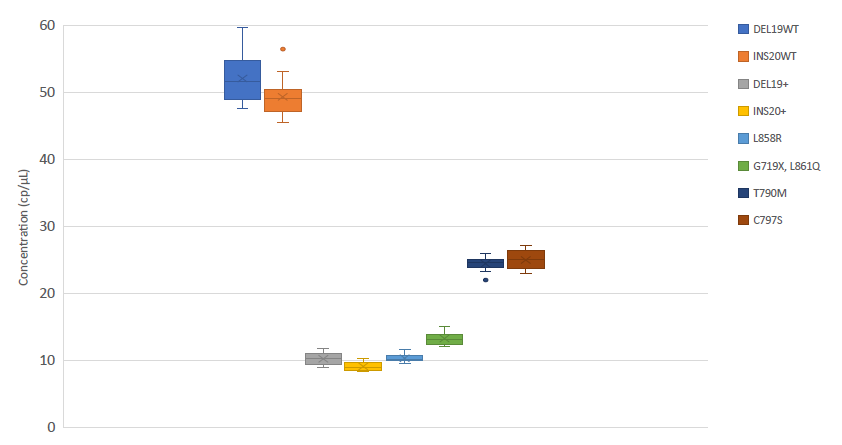
Table 3
Specificity results of Crystal Digital PCR™ using the EGFR 6-color Crystal Digital PCR™ kit. Reactions were performed in duplicate using synthetic Ultramer™ mutant DNA oligonucleotides and wild-type human genomic DNA as templates.
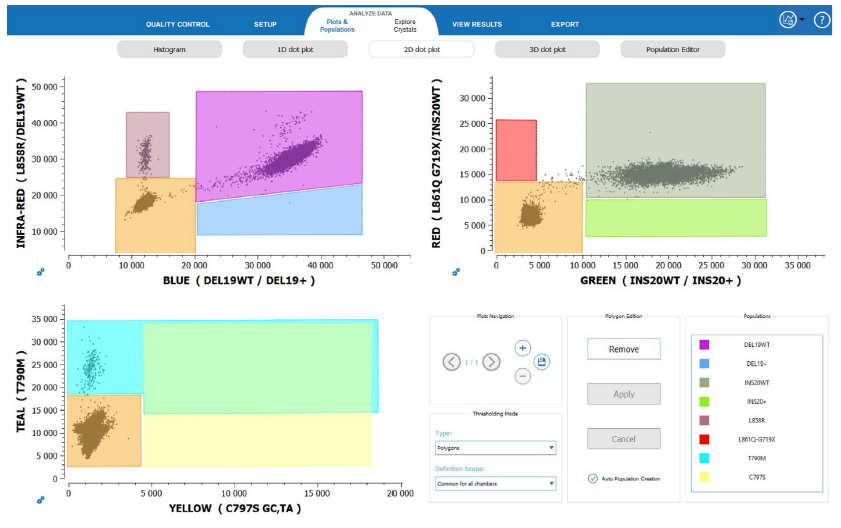
The analysis template provided with the EGFR 6-color Crystal Digital PCR™ kit combined with Crystal Miner software allow for the straightforward generation of 2D-plots (Figure 5) and the rapid obtention of highly concordant results.
Figure 5
A representative example of the analysis of a L858R and T790M positive NSCLC clinical cfDNA sample using Crystal Miner software and the custom EGFR 6-color Crystal Digital PCR™ kit analysis template. The cfDNA sample showed a perfect concordance between the expected and measured results. The x- and y-axis correspond to the fluorescence units of the indicated color channel.
Application Note Highlights
- The EGFR 6-color Crystal Digital PCR™ kit reliably detects 32 common, rare, activating, and resistant somatic EGFR mutations in a single assay.
- The EGFR 6-color Crystal Digital PCR™ kit is optimized for ultrasensitive and highly robust EGFR mutation detection on the naica® system with LoDs ranging from 0.30 to 0.46 cp/μL, depending on the target using Sapphire chips.
- The Crystal Miner software and custom EGFR 6-color analysis template enable fast and straightforward analysis of cfDNA.
To learn more about digital PCR, please visit Stilla Technologies’ Learning Center at www.gene-pi.com