Guidelines for 6-color multiplex assay design for optimized performance with Crystal Digital PCR™
By measuring several targets in a single reaction, multiplex assays enable users to conserve precious sample volume and to save time, reagents and costs. In addition, as all targets are amplified and measured in the same reaction, multiplexing improves quantification precision by reducing pipetting errors that contribute to sample and reagent variations when performing separate single- plex reactions. Multiplexing in Crystal Digital PCR™ is as sensitive and accurate as single-plexing. Careful assay design and assay optimization steps help realize the more complex reaction composition, containing a high number of primers and probes for the amplification of several DNA targets in a single PCR reaction. The Crystal Miner software is an open data analysis software that provides intuitive tools to help optimize and troubleshoot multiplex assays. This Technical Note provides straightforward guidelines to facilitate multiplex assay design using the naica® system.
Guidelines for experimentally evaluating primer and probe performance
- Stilla® recommends using the naica® multiplex PCR MIX, which is specially formulated for optimal multiplexed Crystal Digital PCR™ performance.
- It is imperative to start with a Crystal Digital PCR™ run of the individual reaction composition in the single plex format first. Each primer/probe/target requires a performance verification in the less complex reaction mix before proceeding to multiplexing. For example, for a 6-plex assay, six individual single plex reactions should first be performed on control nucleic acid target templates before combining the reagents in a multiplex reaction. When performing a single-plex reaction, a single positive population is expected.
- For optimized multiplex assay performance, it is important to consider the final sample matrix and composition (e.g., short fragmented DNA for assay design targeting circulating cell-free DNA, more intact DNA fragments for assay design targeting genomic DNA).
- The DNA template used should be devoid of contaminants and potential inhibitors. Synthetic oligos can be used as templates for assay optimization if final sample material is rare or not readily available.
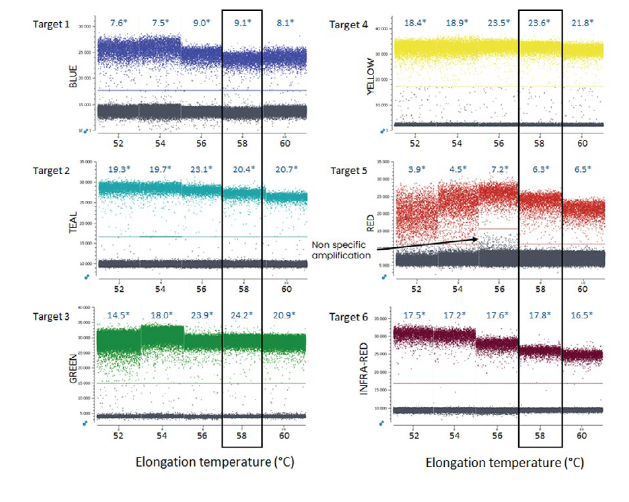
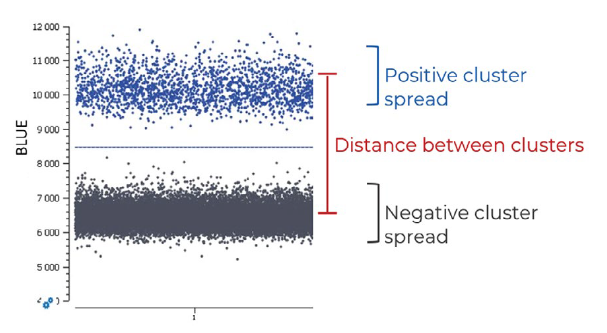
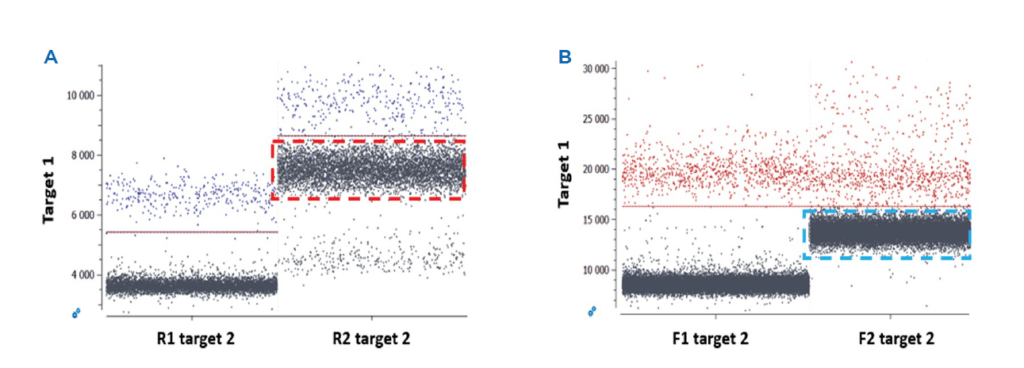
- A range of elongation temperatures should be evaluated for each single-plex reaction to determine the optimal reaction temperature at which there is good separability between positive and negative populations, without nonspecific amplification (Figure 1). The Stilla® separability score provided by Crystal Miner software (Figure 2) should be used as a metric to determine the optimal elongation temperature common to all probes. If the single-plex reaction is not well optimized, a second distinct amplified population may be apparent due to, for example, undesired probe interactions. In addition, non-specific amplification can result from one of several unoptimized parameters, including primer/probe dimers or primer/probe non-specificity. In this case, various methods can be employed to limit the amplification of non-specific sequences, such as increasing the annealing temperature, performing a touchdown PCR or redesigning primer sequences. It is important to evaluate the primer and probe interactions using adapted in silico tools before testing the reagents in vitro.
- Perform multiplex Crystal Digital PCR™ with all primers and probes at the selected elongation temperature and evaluate the reaction performance using the separability score as a guide. If necessary:
- Adjust the number of PCR cycles — It is recommended to start with 45 cycles and to increase the cycle numbers for further optimization of the separability between the positive and negative populations.
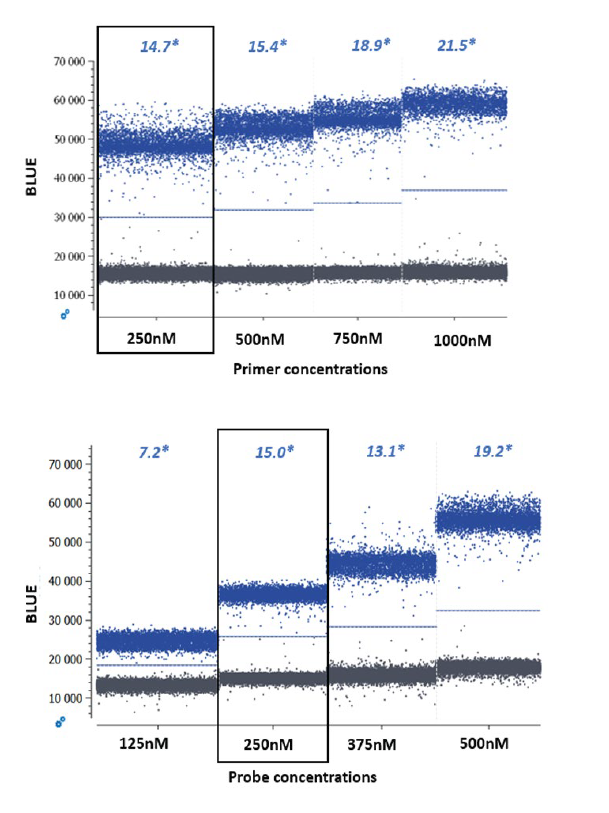
- Adjust primer and probe concentrations — for the naica® system, the recommended primer and probe concentration range from 0.125 to 1 μM (Figure 3). For multiplex assay design it is recommended to start at the low end of the concentration range to minimize the complexity of the reaction and reduce the volume occupied by primers and probes.
- Use modified bases such as locked nucleic acid (LNA)1 bases or a minor groove binder (MGB)2 to increase the Melting temperature (Tm) of the probe while keeping a short length (<20nt if possible). However, no more than 2 MGBs are recommended in a multiplex assay due to a risk of decreased amplification
- Evaluate primer and probe interactions — The probability of homo/hetero dimer formation between primers and/or probes used in the same multiplex experiment should be kept to a minimum. Dimerization can be evaluated, and interaction scores determined with several in silico design tools (e.g., IDT OligoAnalyzer™ Tool, Primer3, Applied Biosystems™ Primer Express®, PREMIER Biosoft Beacon Designer™) (Figure 4). High concentrations of primers and probes can increase the probability of undesired interactions. Thus, when multiplexing, it is recommended to start with low concentrations of primers for all assays (e.g. 0.25 μM), and increase the concentrations gradually up to 1 μM if needed (for example, to increase the amplification efficiency).
- For a multiplex assay, it is important to compensate for fluorescence spillover to ensure robust quantification. Using monocolor controls, the Crystal Miner software allows to create a compensation matrix adapted to a given multiplex panel. Learn more about Fluorescence Spillover. Instructions for performing fluorescence spillover compensation can also be found in the Crystal Miner software User Manual.
Technical Note Highlights
- The naica® system offers flexible 6-color multiplexing capabilities.
- The Crystal Miner software facilitates multiplex assay optimization and troubleshooting.
- The Technical Note provides cost- and time-saving tips to design and validate Crystal Digital PCR™ multiplex assays on the naica® system.
- Designing multiplex assays is not complex if the guidelines provided in this Technical Note are followed
Endnotes
- Locked nucleic acids in PCR primers increase sensitivity and performance. Ballantyne KN, van Oorschot RA, Mitchell RJ. Genomics. 2008 Mar;91(3):301-5. doi: 10.106/j.ygeno.2007.10.016 [PMID: 18164179]
- 3′-Minor groove binder-DNA probes increase sequence specifity at PCR extension temperatures. Kutyavin IV, et al. Nucleic Acids Research. 2000 Jan;28(2): 655 661. doi. org/10.1093/nar/28.2.655.