Detect more targets in a single Crystal Digital PCR™
using amplitude-based multiplexing
Introduction
A multiplex PCR assay is defined as the simultaneous amplification of more than one target in a single reaction. By contrast, the amplification of a single target in a single reaction is referred to as a simplex or single-plex assay. Detection of several targets in multiplex Crystal Digital PCR™ (cdPCR) is as sensitive and accurate as detection in a simplex reaction. There are several benefits of running a multiplex assay compared to running multiple simplex assays. In a multiplex assay, the same results are obtained while conserving precious samples, saving time, reducing reagents and subsequent analysis costs, and reducing pipetting variation. The high number of detection channels of the naica® system (3-color and 6-color), enable users to increase multiplexing capacity, thereby detecting a greater number of targets in a single reaction.
In the simplest example of cdPCR, a single target can be detect- ed in a single fluorescence detection channel. As the number of detection channels increases, the number of targets that can be simultaneously detected in a single reaction also increases. For example, with the 6-color naica® system, if each color channel is used to detect a single target, six targets can be detected in a single reaction (6plex). However, additional strategies can be employed to allow the detection of more than one target per color channel. One such strategy is fluorescence amplitude-based multiplexing. In an amplitude-based multiplex assay, multiple targets are detected and quantified independently in the same color channel by employing multiple dual-labeled fluorescent probes (TaqMan® probes) conjugated with the same fluorescent dye and same quencher. A distinction can be made between each target by, for example, varying the final concentration of each probe within a given color channel such that after end-point amplification, the relative fluorescence intensity value of each target is distinguishable from that of the other target detected in the same channel.
- For the 3-color naica® system this methodology allows to establish up to 6plex assays.
- For the 6-color naica® system this methodology allows to establish up to 12plex assays.
This Technical Note details a set of straightforward steps to create a highly multiplexed cdPCR assay using all detection channels of the 6-color naica® system and fluorescence amplitude-based multiplexing.
Step 1: In silico design of primers and probes
The principles of in silico design of primers and probes for a multiplex assay have been previously described in a step-by- step guide to multiplex Crystal Digital PCR™ design Technical Note for the naica® system, available on the Resources section of the Stilla Technologies webpage.
- 3-color naica® system: “Guidelines for 3-color multiplex assay design for optimized performance with Crystal Digital PCR™”
- 6-color naica® system: “Guidelines for 6-color multiplex assay design for optimized performance with Crystal Digital PCR™”
Stilla Technologies recommends using the naica® multiplex PCR MIX, which is specially formulated for optimal multiplexed Crystal Digital PCR™ performance.
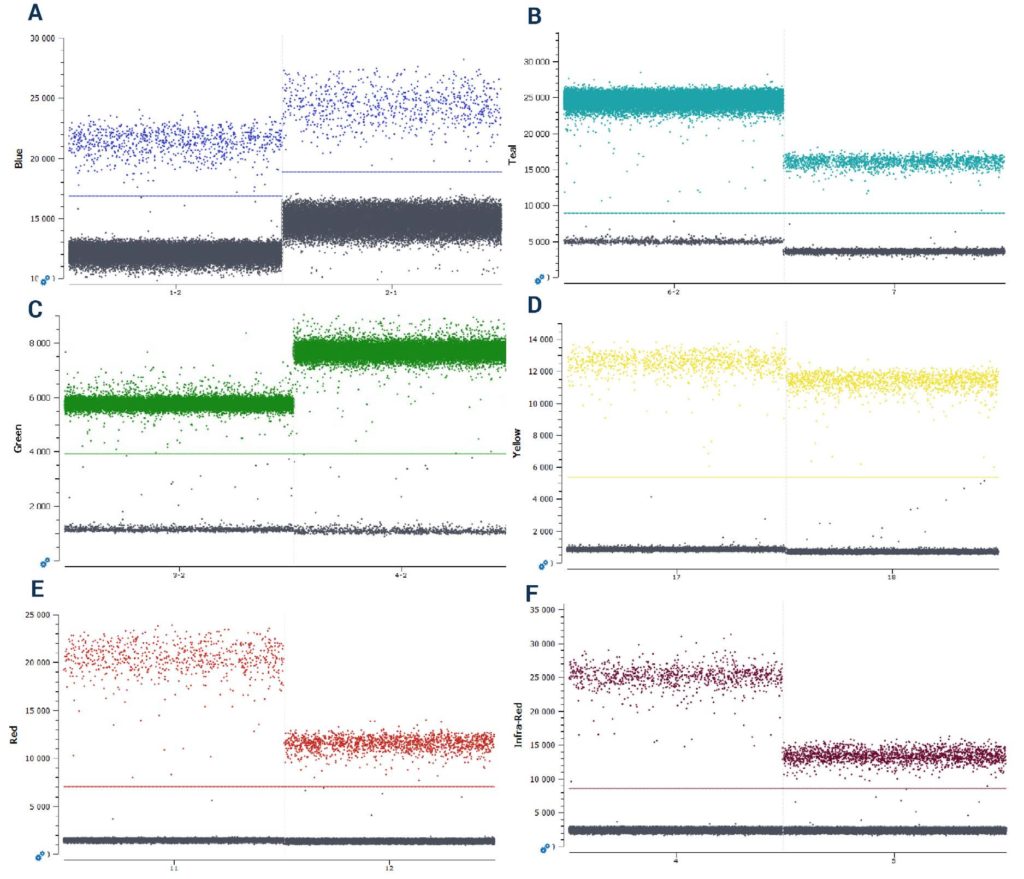
(E) Cy5 (F) Cy5.5.
Step 2: Implementation of a simplex assay
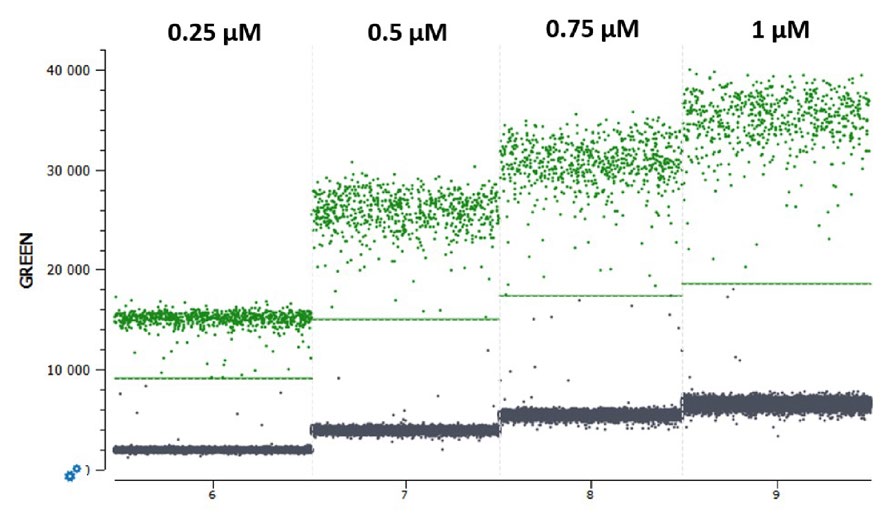
In cdPCR experiments, the final reaction concentration of a fluorescent probe influences the final fluorescence intensity level obtained upon detection of a given target: higher probe concentrations tend to yield higher fluorescence levels (Figure 1). As indicated in the respective naica® system guide to multiplex design Technical Note, the negative and the positive populations of each target must be separated sufficiently to place unambiguously a threshold line (Figure 1).
However, not all probes behave alike. Two probes labeled with the same fluorophore and quencher but composed of two unique sequences can exhibit differences in their final fluorescence intensity levels when used at the same final reaction concentration (Figure 2). Thus, it is crucial to conduct first a simplex experiment with each probe at a fixed concentration (Stilla Technologies recommends starting at a concentration of 0.25 µM in the reaction) to determine a base-line fluorescence level for each probe at that concentration before proceeding to fluorescence amplitude-based multiplexing.
Step 3: Implementation of a duplex assay using fluorescence amplitude-based multiplexing in a single-color channel
Once the fluorescence level of each probe at a given concentration is determined, probes with the same fluorophore can be tested together to evaluate their fluorescence intensities within a multiplexed reaction in the same color channel.
A multiplex experiment is defined by the presence of two or more probes and their corresponding primers in the same reaction mix. To set up an amplitude-based multiplexing reaction, two probes detected in the same fluorescence channel are added together in a single reaction mix at the concentrations defined above in Step 2. Stilla recommends using probes with the same fluorophore and quencher combination for a given color channel to ensure optimal fluorescent spillover compensation and analysis.
Note: For any given color channel, the fluorescence level obtained from the detection of target 1 must be adequately different from the fluorescence level obtained for target 2 and from the double positives. This difference in fluorescence level must allow to distinguish the populations and unambiguously place the polygon thresholds around each population using the Crystal Miner analysis software. If the separability between each population is not adequate to place a threshold, the probe concentrations should be readjusted to obtain three distinct positive populations, allowing straightforward thresholding and robust target quantification.
The localization of the positive populations on the dot-plot depends on several factors. However, the degree of competition between the two probes has a strong influence on the positive droplet cluster position. Two reaction categories can be distinguished:
1. Non-competing duplex reactions (two primers pairs, two probes)
A “classic” duplex reaction generates two amplicons using two distinct primer pairs and generates a fluorescence signal emanating from two distinct probes (Figure 3A). In this scenario, in a 1D dot-plot, the lowest fluorescence intensity positive population is composed of droplets containing only target 1, the middle fluorescence intensity positive population is composed of droplets containing only target 2 and the uppermost population is composed of droplets containing both target 1 and target 2 (Figure 3A).
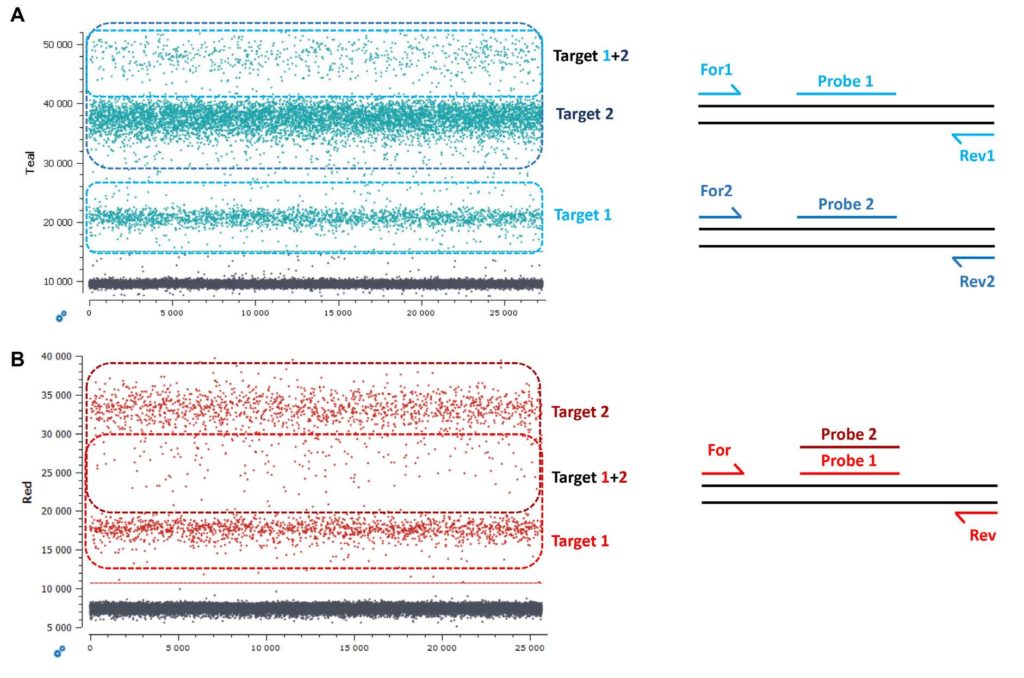
2. Competing duplex reactions (one primer pair with two probes binding to the same sequence region)
Another possibility is to target the same sequence region with two probes amplified with a single primer pair (Figure 3B).
Competing duplex reactions can occur when detecting two mutations that are positioned on the same sequence base (ex. mutations EGFR G719A or EGFR G719S) or when detecting a mutant sequence and its corresponding wildtype sequence (ex. EGFR G719A and wildtype EGFR G719).
Taking the case of EGFR G719A and wildtype EGFR G719, due to the competition between the probes and the possibility to have co-encapsulation of different quantity ratios of wildtype and mutant sequences in a single droplet (ex. 1:1 vs 2:1 vs 1:2), the double positive droplets will be positioned between the two single positive droplet populations on the 1D dot-plot. The double positive droplets can be easily mistaken for rain if the assay is not highly optimized or if the targets are at low concentrations (Figure 3B).
In each type of duplex assay, the three positive populations must be distinct from one another to ensure accurate quantification of each target.
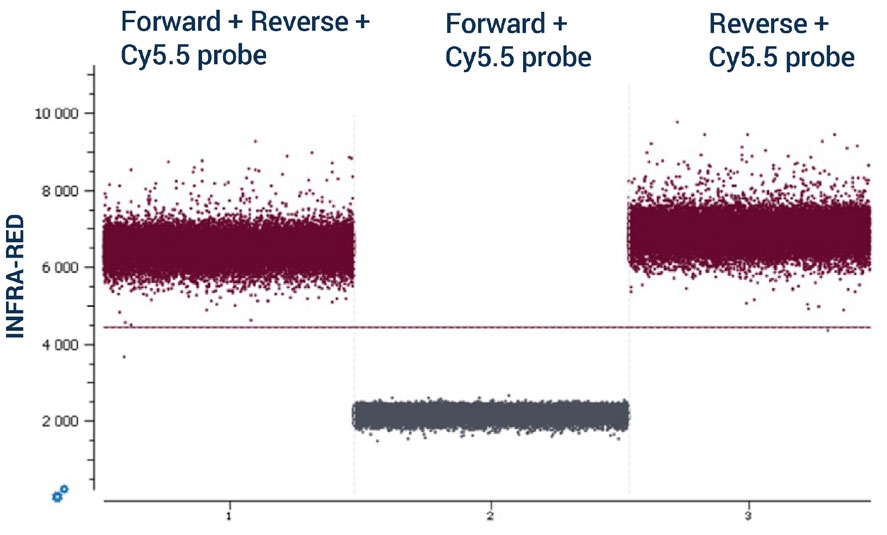
To increase the number of targets detected in a single assay and by following the recommendations detailed in Steps 1, 2, and 3, multiple duplex reactions can be combined using different color channels of the naica® system to yield a highplex assay. For example, a 12plex assay can be designed where two targets are detected in each of the detection channels of the 6-color naica® system. Note: It is mandatory to evaluate the absence of non-specific interactions between primers and probes in silico and to confirm their absence experimentally as non-specific interactions can negatively impact the performance of an assay. Non-specific interactions can occur either between the primers and probes used to detect targets in the same color channel or between the primers and probes used to detect targets in different color channels. To evaluate experimentally the absence of non-specific interactions, Stilla Technologies recommends conducting a non-template control (NTC) cdPCR containing primers and probes but no DNA template. In an NTC, the basal fluorescence in each color channel should be low. However, if non-specific interactions occur between primers and probes, the basal level of fluorescence in the corresponding color channel can be elevated in an NTC reaction. Stilla Technologies recommends starting with simplex NTC reactions and building up to duplex NTC and higher multiplex NTC reactions when evaluating non-specific interactions. In the simplex example in Figure 4, a non-specific interaction occurs between a primer and the probe used to detect a target in the same color channel: the reverse primer of the infrared-detected target and the infrared (Cy5.5) probe. The non-specific interaction was first revealed by the high level of basal fluorescence observed in the simplex NTC reaction where the forward and the reverse primers and infrared Cy5.5 probe were included (Figure 4, chamber 1). By removing either the reverse primer (Figure 4, chamber 2) or the forward primer (Figure 4, chamber 3) of the NTC reaction and comparing their basal fluorescence intensities, the interaction between the reverse primer and the infrared (Cy5.5) probe was identified. To address this non-specific interaction, the reverse primer was redesigned and the simplex NTC experiment repeated to ensure that the non-specific interaction was removed.
If the non-specific interaction occurs between primers and probes of different color channels, the presence of the interaction can be identified by comparing the level of basal fluorescence of the negative droplet population of each simplex NTC reaction to that of the multiplexed NTC reaction. If a substantial increase in the basal fluorescence level is observed in the multiplexed reaction, it suggests the presence of a heterodimer interaction. By removing each primer one by one, the identity of the interaction can be revealed.
If primers and probes display undesired interactions, Stilla Technologies recommends redesigning the problematic primer and redoing the NTC reactions to re-evaluate the interaction.
Once optimal primer and probe sequence and final concentrations are established, the cdPCR program can be further optimized to allow higher multiplexing (Step 4).
Step 4: Compensating fluorescence spillover and increasing multiplexing
Multiplex assay design on the naica® system requires the generation of a compensation matrix to correct for fluorescence spillover from one color channel to another, thus ensuring more accurate target quantification.
A compensation matrix can be easily generated by following the instructions in the Technical Note “How to generate a fluorescence compensation matrix for multiplex assays on the naica® system.” A comprehensive detailed description is also available in the respective naica® system User Manual for Crystal Miner software. For amplitude-based multiplexing, if the same fluorophore and quencher are used for both probes in the color channel, the fluorescence spillover from the two probes into the adjacent color channels should be approximately the same. Thus, for amplitude-based multiplexing, Stilla Technologies recommends adding only one of the two target DNA templates (the template corresponding to the highest intensity probe) for each monocolor control reaction rather than adding both target DNA templates.
Like lower-plexed cdPCR assays, amplitude-based highplex cdPCR assays can be ultrasensitive, with the limit of blank (LoB) of each target approaching zero in a 12plex assay (Table 1). To calculate the LoB of an assay, Stilla Technologies recommends following the method outlined in the Technical Note “How to characterize the Limit of Blank and the Limit of Detection in Crystal Digital PCR™.”
| Target | LoB copies/µL |
| Blue 1 | 0.00 |
| Blue 2 | 0.09 |
| Teal 1 | 0.00 |
| Teal 2 | 0.08 |
| Green 1 | 0.07 |
| Green 2 (Reference gene) | NA |
| Yellow 1 | 0.00 |
| Yellow 2 | 0.08 |
| Red 1 | 0.06 |
| Red 2 | 0.15 |
| InfraRed 1 | 0.00 |
| InfraRed 2 | 0.06 |
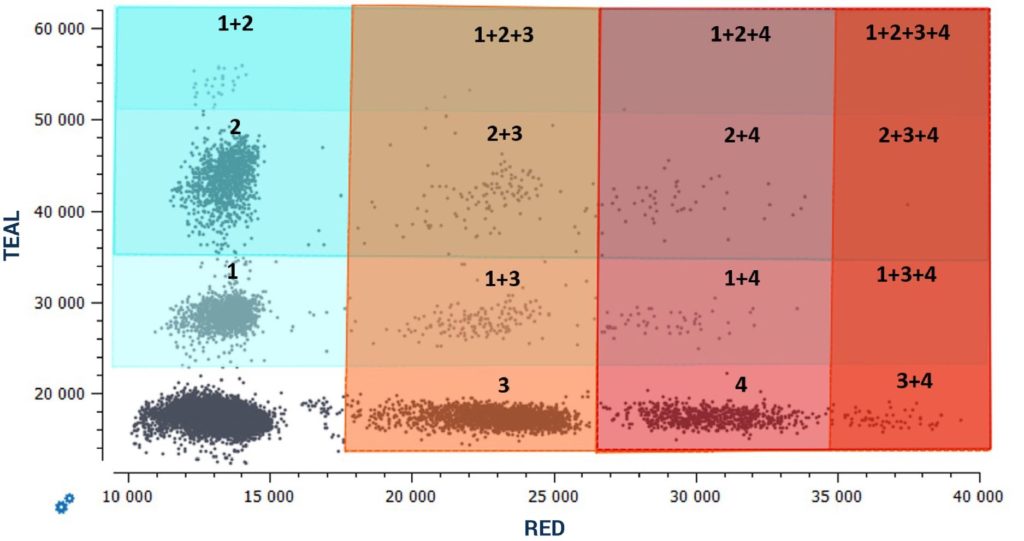
By following the provided guidelines to create a highly multiplexed cdPCR assay using all detection channels of the naica® system and fluorescence amplitude-based multiplexing, highly sensitive 6plex multiplex assays detecting up to six unique targets in a single reaction can be established on the 3-color naica® system and highly sensitive 12plex multiplex assays de- tecting up to twelve unique targets in a single reaction can be established on the 6-color naica® system (Figure 5).
Technical Note Highlights
- The 3-color naica® system and 6-color naica® system offer flexible highly sensitive multiplexing capabilities to increase the number of targets detected in a single assay
- Using amplitude-based multiplexing of two targets per color channel, the number of targets detected can be doubled, reaching up to six targets for a 3-color naica® system and up to twelve targets for a 6-color naica® system
- Amplitude-based multiplexing increases the quantity of information obtained from a single reaction without compromising the assay’s sensitivity, maximizing the use of any precious sample
Learn more about what the Nio™+ instrument and Crystal Digital PCR™ can do for your research