Effect of Inhibitors – Crystal Digital PCR™ vs qPCR
Effect of inhibitors in PCR-based quantification
There are thousands of chemical species that can act as inhibitors in PCR-based reactions. These compounds, present in the initial sample, may be co-purified with DNA and can be hard to get rid of.
Quantification using real-time PCR (qPCR) versus Crystal Digital PCR
Real-time PCR quantification is based on comparing Cq values between unknown samples and standards containing known amounts of DNA. For adequate quantification, this technique therefore relies on similar PCR efficiencies between the analyzed samples and the standards. However, in presence of an inhibitory concentration of a given substance in the sample, PCR efficiency may differ between standard and sample, thus biasing quantification (Figure IA). In Crystal Digital PCR, the droplets are characterized as either positive or negative, whereas the amplitude of fluorescence separating the two populations indicates PCR efficiency. Even in case of decreased PCR efficiency due to inhibitors, accurate quantification can still be achieved (Figure 1B).

Figure 1. Effect of humic acid in real-time PCR (A) and Crystal Digital PCR (B). PCR reactions for either qPCR or Crystal digital PCR all contained IX PerfeCta Multiplex qPCR ToughU-mix, 100 nM fluorescein, 500nM of forward and reverse primers for the ALB gene, 250 nM of Cy-5 labelled hydrolysis probe to detect the amplicons and 5.4 ng/uL of human genomic DNA. These reactions were spiked with either O, 50 or 100 pg/mL of humic acid.
It is also important to bear in mind that the presence of inhibitors may diversely affect quantification of different targets from the same source DNA (Figure 2). Taking advantage of the 3-color capabilities of the Naica™ system can help you target different regions on the gene or genome of interest to check for such bias.
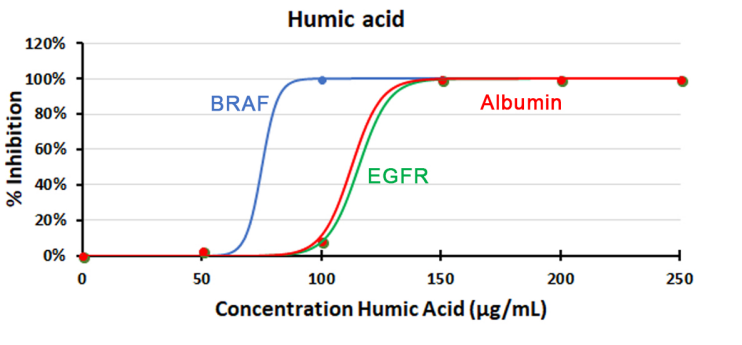
Figure 2. Effect of humic acid inhibitor in Crystal Digital PCR on three different targets. Multiplex Crystal digital PCR reactions containing PerfeCta Multiplex qPCR Toughmix, primers and probes targeting the ALB gene (detected with Cy-5-labelled probe), EGFR gene (detected with HEX-Iabelled probe) and BRAF (detected with FAM-labelled probe), and identical quantities of human genomic DNA were spiked with increasing concentrations of humic acid (plotted with sigmoid fit). Percentage of inhibition is calculated considering 0% inhibition for DNA quantification with no inhibitor.
Comparing tolerance of qPCR and Crystal Digital PCR™ to inhibitors
We compared the effects of two known inhibitors, humic acid, found in soil samples, and heparin, used as an anticoagulant in collected blood samples, on DNA quantification both in real-time PCR and Crystal Digital PCR. We found that accurate quantification remains possible in presence of higher concentrations of either inhibitor when using Crystal Digital PCR, compared to real-time PCR (Figure 3).
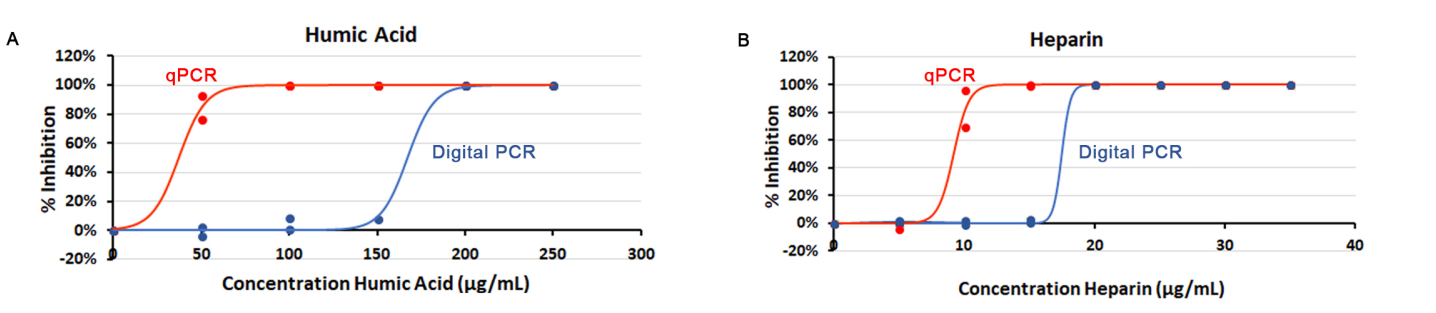
Figure 3. Concentration-dependent inhibitory effect of humic acid (A) and heparin (B) on real-time PCR (qPCR) and Crystal Digital PCR. PCR reactions for either qPCR or Crystal digital PCR contained PerfeCta Multiplex qPCR Toughmix, identical quantities of human genomic DNA, as well as primers and probes targeting either the BRAF gene when testing for humic acid or EGFR gene when testing for heparin. All samples were assayed in triplicates for N=2 experiments (plotted with sigmoid fit). Percentage of inhibition is calculated considering 0% inhibition for DNA quantification with no inhibitor.
In conclusion, when dealing with inhibition-prone samples, digital PCR can be more reliable than qPCR.
It may just be the solution you have been looking for!