Detection of HER2 Copy Number Variation with Crystal Digital PCR™
Triplex Crystal Digital PCR for HER2 Copy Number amplification
Investigating HER2 amplification status is crucial in breast cancer to determine the indication of anti-HER2 targeted therapy. Assessing HER2 copy number variation is challenging especially when tumor DNA is diluted in DNA from healthy cells. Here, we illustrate how Crystal Digital PCR is capable of reliably identifying HER2 amplification in samples with low tumoral fraction.
An assay previously described for the detection of HER2 (ERBB2) and MRMI on chromosome 17, and TSN (reference gene) on chromosome 2[1], was optimized for Crystal Digital PCR. This assay enables the quantification of HER2 copy number and the distinction between HER2 amplification and chromosome 17 polysomy, a condition that could lead to HER2 status misinterpretation.
Evaluating sensitivity of HER2 amplification detection
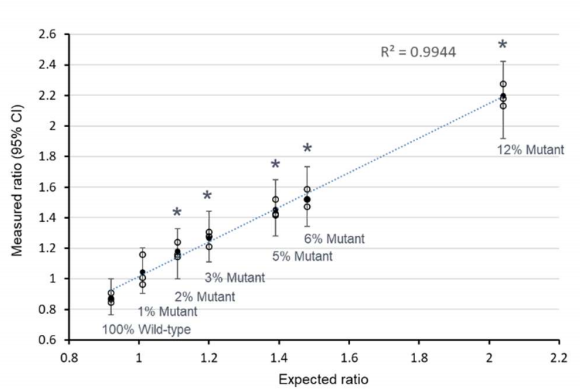
Genomic DNA extracted from SKBR3 cell line (HER2/TSN ratio of 10:1) was spiked into non-amplified genomic DNA with mutant fractions from 1% to 12%, modeling a theoretical range of HER2/TSN ratios from 1 to 2.2.
Absolute concentrations for TSN and HER2 were calculated using Poisson law and a Z-test was applied on the log-ratio HER2/TSN (approximated by a Normal distribution) to interrogate the presence of mutant DNA with statistical significance of α=β=5%[2]. Using this method, we were able to significantly detect the presence of mutant DNA for mutant fractions as low as 2%, corresponding to a HER2/TSN normalized ratio of 1.2.
HER2 amplification and chromosome 17 polysomy status
Current methods to evaluate HER2 amplification in breast tumor samples rely on immunohistochemistry (IHC) to assess HER2 overexpression, which, for equivocal IHC HER2 expression (IHC2+), may be complemented by fluorescence in-situ hybridization (FISH) to detect HER2 gene amplification. These methods are labor-intensive and require a high level of expertise for analysis. Molecular methods may provide simpler, faster and unbiased alternatives.
The present study was conducted using DNA extracted from tumors from 7 patients with breast cancer, provided by Institut Gustave Roussy (France)§, and results obtained by Crystal Digital PCR were compared to those obtained using standard diagnostic procedure.
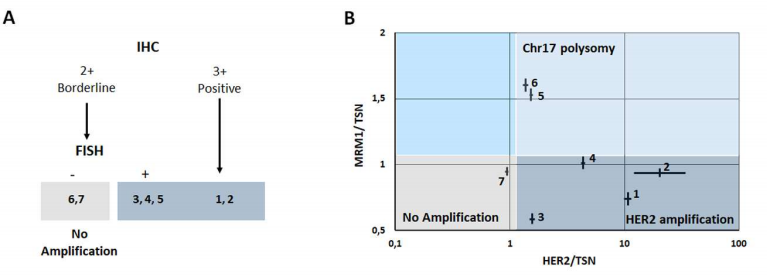
Results obtained with IHC/FISH and Crystal Digital PCR were in agreement for patients 1-4 samples, deemed positive for HER2 amplification by both methods, as well as for samples from patients 6 and 7 identified as negative for HER2 amplification. Crystal Digital PCR™ additionally identified a case of chromosome 17 polysomy for patient 6. However, whereas HER2 amplification was identified for patient 5 sample using standard diagnostic procedure, Crystal Digital PCR identified the elevated HER2 signal as attributable to chromosome 17 polysomy.
[I] Jacquemier et al. “SISH/CISH or qPCR as alternative techniques to FISH for determination of HER2 amplification status on breast tumors core needle biopsies:a multicenter experience based on 840 cases”. BMC Cancer. 2013 ; 13:351. PMID: 23875536
[2] Whale et al. “Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation”. Nucleic Acids Res. 2012 ;40:11. PMID: 22373922
§ Courtesy of Dr Cécile Jovelet, Translational Research Laboratory, Institut Gustave Roussy, France