3-Color Crystal Digital PCR™ assays for EGFR mutation detection
Two multiplex digital PCR assays detect EGFR activating and resistance
In non-small cell lung cancer, the epidermal growth factor receptor (EGFR) is a common therapeutic target. EGFR activating mutations, such as L858R, L861Q, and exon 19 deletions are predictive of disease responsiveness to targeted therapy using tyrosine kinase inhibitors (TKls) [1]. On the contrary, the presence of EGFR T790M mutation is associated with tumor resistance to TKIs [2].
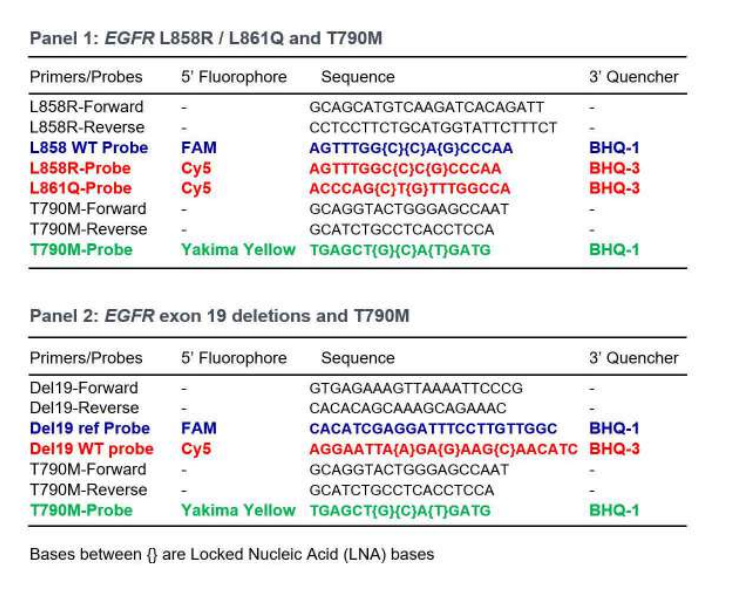
To detect and quantify these mutations in single tests without sacrificing the precision and reliability of the results, two multiplex assays were developed. Primers and probes using three different fluorescence channels of the Naica™ System enable detection of EGFR L858R/L861Q and T790M in panel 1, and EGFR exon 19 deletions (as a drop-off assay) and T790M in panel 2. Both panels also detect wild-type (WT) EGFR.
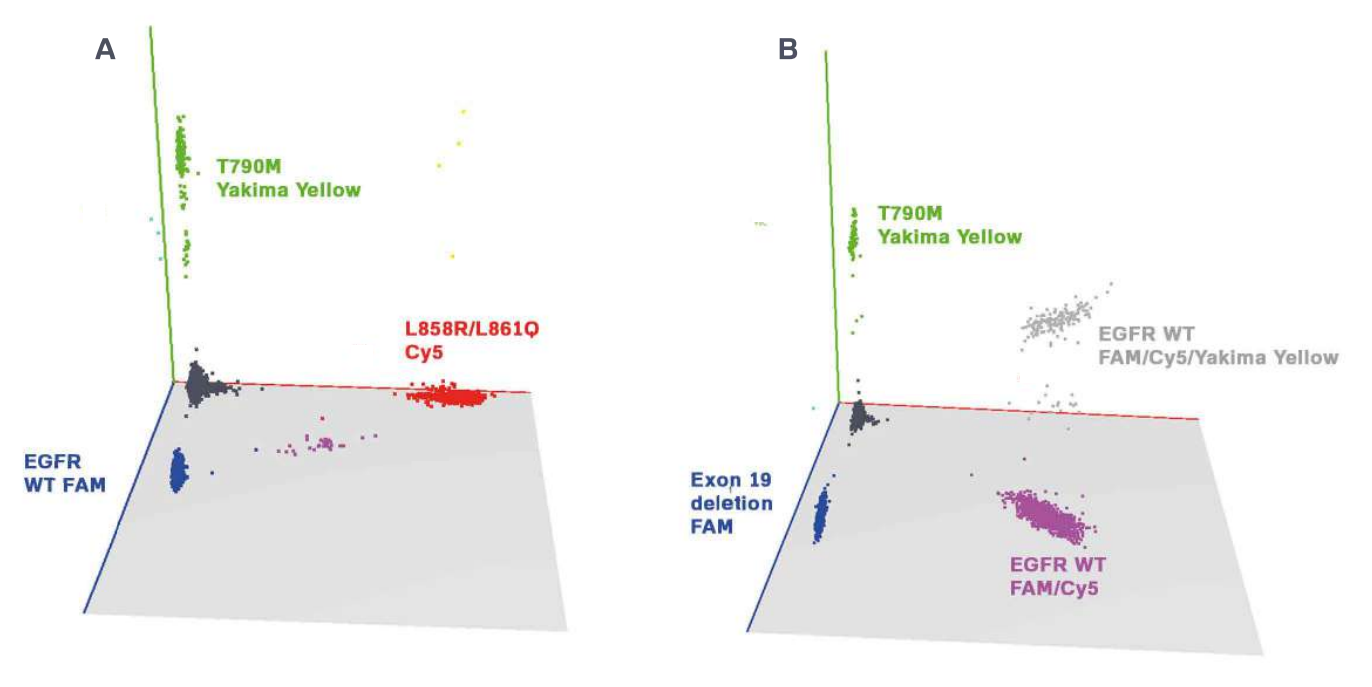
Three-color assays robustly and reliably detect EGFR mutations
The targeted EGFR mutations were detected with a 95% confidence interval at final concentrations down to 1 and 0.4 copies per microliter (cp/µl), depending on the detected mutation. All assays were performed in a background of 400 copies per microliter of wild-type DNA, and thus the EGFR concentrations represent a mutant allele frequency of 0.25% and 0.1%, respectively (Figure 2).
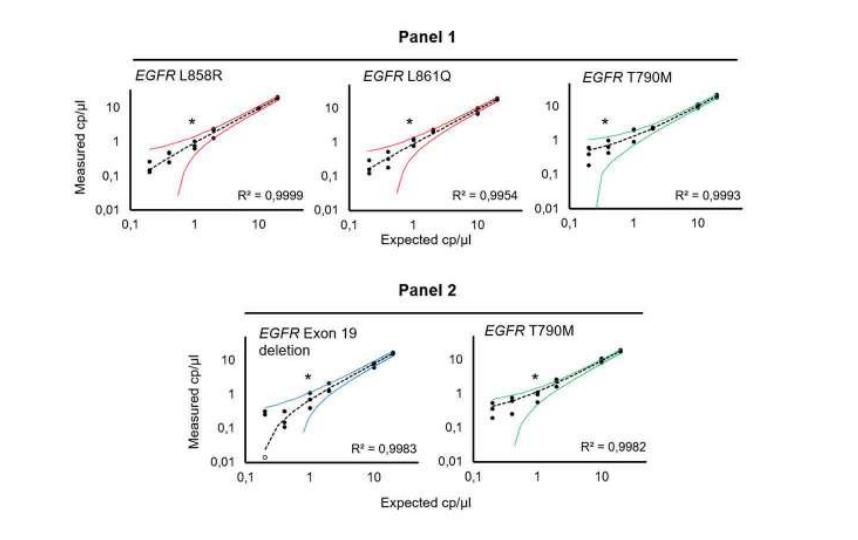
Figure 2: Evaluation of multiplex Crystal Digital PCR performance tor the EGFR multiplex assays. Titration (20 – 0.20 cp/µl) of EGFR L858R, EGFR L861Q, EGFR T790M, and EGFR exon 19 deletion, in a background of 400 cp/µl of WT EGFR (10,000 copies per 25µl reaction). The theoretical 95% confidence intervals are represented as colored curves. *Indicates the theoretical limit of detection at a 95% confidence level interval, derived from the limit of blank measured on 30 replicates containing 10,000 copies of wild-type DNA per 25µl reaction. To learn more about how to determine a limit of blank and a limit of detection, please visit our learning center at www.gene-pi.com
EGFR mutation detection in non-small cell lung cancer patient samples
The two EGFR 3-color digital PCR panels were evaluated on mutant DNA extracted from non-small cell lung cancer patients and compared to next generation sequencing (NGS) measurements. The measured mutant allelic EGFR fractions displayed a strong correlation (Figure 3).

Figure 3: Mutant allele fraction (MAF) of activating (e) and resistance (A) EGFR mutations in DNA extracted from frozen (green: and FFPE (blue) tumor and plasma (red) measured by digital PCR using EGFR panels 1 and 2 and by NGS. Compared samples dis. played a good correlation with a significant Pearson coefficient (R = 0.9746; P < 0.05).
[1] Nan X, Xie C, Yu X, Liu J. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget. 2017 Aug. https://www.ncbi.nlm.nih.gov/pubmed/29088904[2] Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA,Ladanyi M, Riely GO. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGF-R-mutant lung cancers Apr. https://www.ncbi.nlm.nih.gov/pubmed/23470965
Application Note Highlights
- The naica® 3-color Crystal Digital PCR™ system robustly and reliably quantifies EGFR resistant and activating mutations.
- EGFR mutations were detected in a background of wild-type DNA with a 95% confidence level at 0.25% to 0.1% mutant allele frequencies depending on the detected EGFR mutation.
- An excellent correlation was observed between the expected and measured EGFR target concentrations in serial dilution experiments.
- Mutant allelic EGFR fractions measured by 3-color digital PCR and Next Generation Sequencing in tumor and plasma from non-small cell lung cancer patients displayed a strong correlation with a significant Pearson coefficient R of 0.97 (P < 0.05).
To learn more about digital PCR, please visit Stilla Technologies’ Learning Center.